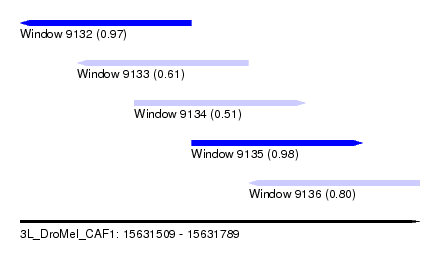
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,631,509 – 15,631,789 |
| Length | 280 |
| Max. P | 0.976943 |

| Location | 15,631,509 – 15,631,629 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.92 |
| Mean single sequence MFE | -41.41 |
| Consensus MFE | -39.58 |
| Energy contribution | -39.83 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15631509 120 - 23771897 AUUUGAAAGGAUAUGUGGCGAGGCAGCCAAAGGACAAUGCCUGGCCGCAACAUAUGCGAAAGGGAUAAUAAUUCACUGGCCGAAAGGCGAUGGGAACAAUGGUCGCAUAACUCAGGCGCG .(((((.......((((((.(((((.((...))....))))).))))))...(((((((.((((((....)))).)).(((....)))..((....))....)))))))..))))).... ( -41.40) >DroSec_CAF1 53758 120 - 1 AUUUGAAAGGAUAUGUGGCGAGGCAGCCAAAGGACAAUGCCUGGCCGCAACAUAUGCGAAAGGGAUAAUAAUUCACUGGCCGAAAGGCGAUGGGAACAAUGGUCGCAUAACUCAGGCGCG .(((((.......((((((.(((((.((...))....))))).))))))...(((((((.((((((....)))).)).(((....)))..((....))....)))))))..))))).... ( -41.40) >DroSim_CAF1 54715 120 - 1 AUUUGAAAGGAUAUGUGGCGAGGCAGCCAAAGGACAAUGCCUGGCCGCAACAUAUGCGAAAGGGAUAAUAAUUCACUGGCCGAAAGGCGAUGGGAACAAUGGUCGCAUAACUCAGGCGCG .(((((.......((((((.(((((.((...))....))))).))))))...(((((((.((((((....)))).)).(((....)))..((....))....)))))))..))))).... ( -41.40) >DroYak_CAF1 58392 120 - 1 AUUUGAAAGGAUUUGUGGCGAGGCAGCCGAAGGACAAUGCCGCGCCGCAACAUAUGCGAAAGGGAUAAUAACUCACUGGCCGAAAGGCGAUGGGAACAAUGGUCGCAUAACUCAGGCGCG .(((((......((((((((.((((..(....)....)))).))))))))..(((((((............((((.(.(((....))).)))))........)))))))..))))).... ( -41.45) >consensus AUUUGAAAGGAUAUGUGGCGAGGCAGCCAAAGGACAAUGCCUGGCCGCAACAUAUGCGAAAGGGAUAAUAAUUCACUGGCCGAAAGGCGAUGGGAACAAUGGUCGCAUAACUCAGGCGCG .(((((.......((((((.(((((.((...))....))))).))))))...(((((((...................(((....)))..((....))....)))))))..))))).... (-39.58 = -39.83 + 0.25)

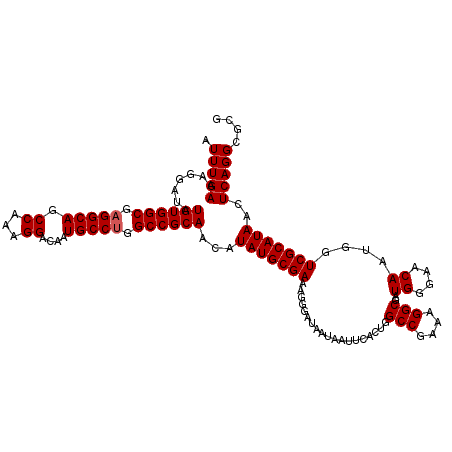
| Location | 15,631,549 – 15,631,669 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -33.78 |
| Energy contribution | -34.27 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15631549 120 - 23771897 GCAAAACGCCCAAUUUCCAAGGCGUGUCCGAGCAACAGACAUUUGAAAGGAUAUGUGGCGAGGCAGCCAAAGGACAAUGCCUGGCCGCAACAUAUGCGAAAGGGAUAAUAAUUCACUGGC (((..(((((..........)))))((((...(((.......)))...)))).((((((.(((((.((...))....))))).)))))).....)))(..((((((....)))).))..) ( -36.10) >DroSec_CAF1 53798 120 - 1 GCAAAACGCCCUAUUUCCAAGGCGUGUCCGAGCAACAGACAUUUGAAAGGAUAUGUGGCGAGGCAGCCAAAGGACAAUGCCUGGCCGCAACAUAUGCGAAAGGGAUAAUAAUUCACUGGC (((..(((((..........)))))((((...(((.......)))...)))).((((((.(((((.((...))....))))).)))))).....)))(..((((((....)))).))..) ( -36.10) >DroSim_CAF1 54755 120 - 1 GCAAAACGCCCAAUUUCCAAGGCGUGUCCGAGCAACAGACAUUUGAAAGGAUAUGUGGCGAGGCAGCCAAAGGACAAUGCCUGGCCGCAACAUAUGCGAAAGGGAUAAUAAUUCACUGGC (((..(((((..........)))))((((...(((.......)))...)))).((((((.(((((.((...))....))))).)))))).....)))(..((((((....)))).))..) ( -36.10) >DroYak_CAF1 58432 120 - 1 GCAAAACGCCCAAUUUUCAAGGCGUGUCCGAGCAACAGACAUUUGAAAGGAUUUGUGGCGAGGCAGCCGAAGGACAAUGCCGCGCCGCAACAUAUGCGAAAGGGAUAAUAACUCACUGGC (((..(((((..........)))))((((...(((.......)))...))))((((((((.((((..(....)....)))).))))))))....)))....(((.......)))...... ( -35.20) >consensus GCAAAACGCCCAAUUUCCAAGGCGUGUCCGAGCAACAGACAUUUGAAAGGAUAUGUGGCGAGGCAGCCAAAGGACAAUGCCUGGCCGCAACAUAUGCGAAAGGGAUAAUAAUUCACUGGC (((..(((((..........)))))((((...(((.......)))...)))).((((((.(((((.((...))....))))).)))))).....)))(..((((((....)))).))..) (-33.78 = -34.27 + 0.50)

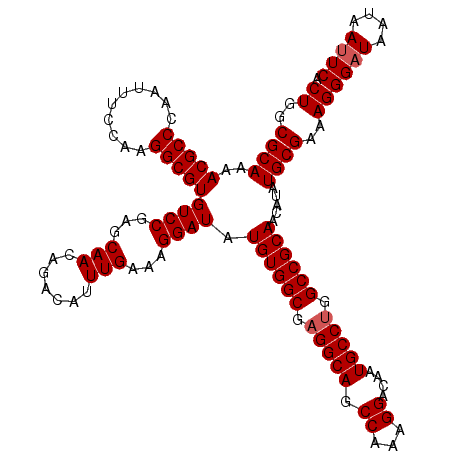
| Location | 15,631,589 – 15,631,709 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -39.80 |
| Consensus MFE | -37.46 |
| Energy contribution | -37.34 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.94 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.505753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15631589 120 + 23771897 GCAUUGUCCUUUGGCUGCCUCGCCACAUAUCCUUUCAAAUGUCUGUUGCUCGGACACGCCUUGGAAAUUGGGCGUUUUGCAGUUAAACGUCGUUGCUGCAAAGUUUUUGGGGCGUGUUCG (((.........(((......)))((((..........))))....))).(((((((((((..((((((.(((((((.......)))))))((....))..))))))..))))))))))) ( -39.70) >DroSec_CAF1 53838 120 + 1 GCAUUGUCCUUUGGCUGCCUCGCCACAUAUCCUUUCAAAUGUCUGUUGCUCGGACACGCCUUGGAAAUAGGGCGUUUUGCAGUUAAACGUCGUUGCUGCAAAGUUUUUGGGGCGUGUUCG (((.........(((......)))((((..........))))....))).(((((((((((..(((((.......((((((((...........)))))))))))))..))))))))))) ( -39.11) >DroSim_CAF1 54795 120 + 1 GCAUUGUCCUUUGGCUGCCUCGCCACAUAUCCUUUCAAAUGUCUGUUGCUCGGACACGCCUUGGAAAUUGGGCGUUUUGCAGUUAAACGUCGUUGCUGCAAAGUUUUUGGGGCGUGUUCG (((.........(((......)))((((..........))))....))).(((((((((((..((((((.(((((((.......)))))))((....))..))))))..))))))))))) ( -39.70) >DroYak_CAF1 58472 120 + 1 GCAUUGUCCUUCGGCUGCCUCGCCACAAAUCCUUUCAAAUGUCUGUUGCUCGGACACGCCUUGAAAAUUGGGCGUUUUGCAGUUAAACGUCGUGGCUGCAAAGUUUUCGGGGCGUGUUCG (((.........(((......)))...........((......)).))).((((((((((((((((((.......((((((((((.......)))))))))))))))))))))))))))) ( -40.71) >consensus GCAUUGUCCUUUGGCUGCCUCGCCACAUAUCCUUUCAAAUGUCUGUUGCUCGGACACGCCUUGGAAAUUGGGCGUUUUGCAGUUAAACGUCGUUGCUGCAAAGUUUUUGGGGCGUGUUCG (((.........(((......)))((((..........))))....))).((((((((((((((((((.......((((((((...........)))))))))))))))))))))))))) (-37.46 = -37.34 + -0.12)

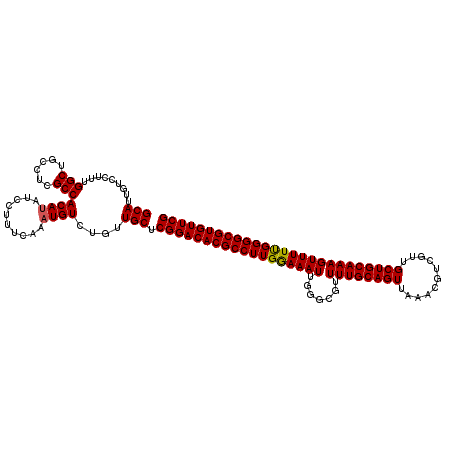
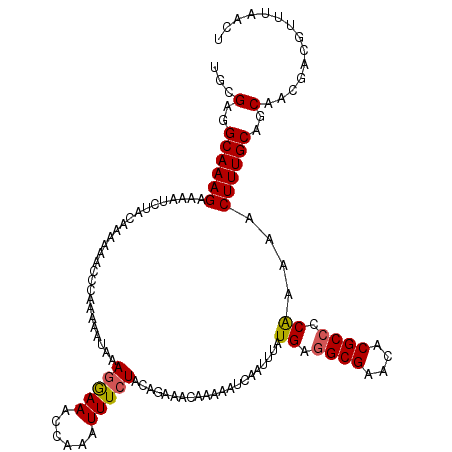
| Location | 15,631,629 – 15,631,749 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -39.23 |
| Consensus MFE | -36.78 |
| Energy contribution | -36.65 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15631629 120 + 23771897 GUCUGUUGCUCGGACACGCCUUGGAAAUUGGGCGUUUUGCAGUUAAACGUCGUUGCUGCAAAGUUUUUGGGGCGUGUUCGCCUCAUAAAUUGAUUUUUGUUUCUGUAGAAAUUUGGUUUC (((((.....)))))((((((........)))))).(((((((...........)))))))(((((.(((((((....))))))).)))))((((...(((((....)))))..)))).. ( -37.90) >DroSec_CAF1 53878 120 + 1 GUCUGUUGCUCGGACACGCCUUGGAAAUAGGGCGUUUUGCAGUUAAACGUCGUUGCUGCAAAGUUUUUGGGGCGUGUUCGCCUCAUAAAUUGAUUUUUGUUUCUGUAGAAAUUUGGUUUC (((((.....)))))((((((((....)))))))).(((((((...........)))))))(((((.(((((((....))))))).)))))((((...(((((....)))))..)))).. ( -39.60) >DroSim_CAF1 54835 120 + 1 GUCUGUUGCUCGGACACGCCUUGGAAAUUGGGCGUUUUGCAGUUAAACGUCGUUGCUGCAAAGUUUUUGGGGCGUGUUCGCCUCAUAAAUUGAUUUUUGUUUCUGUAGAAAUUUGGUUUC (((((.....)))))((((((........)))))).(((((((...........)))))))(((((.(((((((....))))))).)))))((((...(((((....)))))..)))).. ( -37.90) >DroYak_CAF1 58512 120 + 1 GUCUGUUGCUCGGACACGCCUUGAAAAUUGGGCGUUUUGCAGUUAAACGUCGUGGCUGCAAAGUUUUCGGGGCGUGUUCGCUUCAUAAAUUGAUUUCUGGUUCUGUAGAAAUUUGGUUUU ..........((((((((((((((((((.......((((((((((.......))))))))))))))))))))))))))))......((((..(((((((......)))))).)..)))). ( -41.51) >consensus GUCUGUUGCUCGGACACGCCUUGGAAAUUGGGCGUUUUGCAGUUAAACGUCGUUGCUGCAAAGUUUUUGGGGCGUGUUCGCCUCAUAAAUUGAUUUUUGUUUCUGUAGAAAUUUGGUUUC (((((.....)))))((((((........)))))).(((((((...........)))))))(((((.(((((((....))))))).)))))((((((((......))))))))....... (-36.78 = -36.65 + -0.13)


| Location | 15,631,669 – 15,631,789 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -17.62 |
| Consensus MFE | -14.70 |
| Energy contribution | -14.83 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15631669 120 - 23771897 UUCGAGGCAAAGAAAAUCUACAAAAAACCCAAAAAUAAAGGAAACCAAAUUUCUACAGAAACAAAAAUCAAUUUAUGAGGCGAACACGCCCCAAAAACUUUGCAGCAACGACGUUUAACU ...(..((((((...........................(((((.....))))).....................((.((((....)))).))....))))))..).............. ( -17.40) >DroSec_CAF1 53918 120 - 1 UGCGAGGCAAAGAAAAUCUACAAAAAACCCAAAAAUAAAGGAAACCAAAUUUCUACAGAAACAAAAAUCAAUUUAUGAGGCGAACACGCCCCAAAAACUUUGCAGCAACGACGUUUAACU (((...((((((...........................(((((.....))))).....................((.((((....)))).))....)))))).)))............. ( -20.00) >DroSim_CAF1 54875 120 - 1 UGCGAAGCAAAGAAAAUCUACAAAAAACCCAAAAAUAAAAGAAACCAAAUUUCUACAGAAACAAAAAUCAAUUUAUGAGGCGAACACGCCCCAAAAACUUUGCAGCAACGACGUUUAACU (((...((((((...........................(((((.....))))).....................((.((((....)))).))....)))))).)))............. ( -18.90) >DroYak_CAF1 58552 119 - 1 UUCGAGGCAAAGAAAAUCUACAAA-AACCCAAAAAUAAAGAAAACCAAAUUUCUACAGAACCAGAAAUCAAUUUAUGAAGCGAACACGCCCCGAAAACUUUGCAGCCACGACGUUUAACU .(((.(((..((.....)).....-..........(((((........((((((........))))))...........(((....)))........)))))..))).)))......... ( -14.20) >consensus UGCGAGGCAAAGAAAAUCUACAAAAAACCCAAAAAUAAAGGAAACCAAAUUUCUACAGAAACAAAAAUCAAUUUAUGAGGCGAACACGCCCCAAAAACUUUGCAGCAACGACGUUUAACU ...(..((((((..........................(((((......))))).....................((.((((....)))).))....))))))..).............. (-14.70 = -14.83 + 0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:59 2006