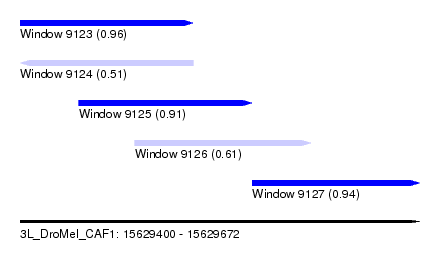
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,629,400 – 15,629,672 |
| Length | 272 |
| Max. P | 0.958344 |

| Location | 15,629,400 – 15,629,518 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.84 |
| Mean single sequence MFE | -24.55 |
| Consensus MFE | -21.99 |
| Energy contribution | -22.05 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15629400 118 + 23771897 ACCGGAGCAACCCGAUUCCAAAAAUCCAAAAAUCCAGAGAUCCAUA--GAUCCAGAGUAAAGCAGCCAGGCUGAACGGAACCAACAUUACUCAAUGUCAACAUCAAAAUUGCUUUGGAAU .(((((((((...(.((((.................(.((((....--)))))...((..(((......)))..)))))).).(((((....)))))...........)))))))))... ( -24.70) >DroSec_CAF1 51628 118 + 1 ACCGGAGCAACCCGACUUCAAAAAUCCCAAAAUCCAGAGAUCCAUA--GAUCCAGAGCAAAGCAGCCAGGCUGAACGGAACCAACAUUACUCAAUGUCAACAUCAAAAUUGCUUUGGAAU .(((((((((.(((......................(.((((....--))))).........((((...))))..))).....(((((....)))))...........)))))))))... ( -23.20) >DroSim_CAF1 52637 118 + 1 ACCGGAGCAACCCGACUCCAAAAAUCCAAAAAUCCUAAGAUCCAUA--GAUCCAGAGCAAAGCAGCCAGGCUGAACGGAACCAACAUUACUCAAUGUCAACAUCAAAAUUGCUUUGGAAU ...((((........))))...........................--..((((((((((..((((...))))..........(((((....)))))...........)))))))))).. ( -24.20) >DroYak_CAF1 56196 120 + 1 ACCGAAGCAACCCGACUCCAAAAAUCCACAGAUCUAAAAAUCCAUAGAGAUCCAGAGUAAAGCAGCCAGGCUGAACGGAAGCAACAUUACUCAAUGUCAACAUCAAAAUUGCUUUGGAAU .(((((((((....................(((((............)))))..((((((..((((...))))..(....).....))))))................)))))))))... ( -26.10) >consensus ACCGGAGCAACCCGACUCCAAAAAUCCAAAAAUCCAAAGAUCCAUA__GAUCCAGAGCAAAGCAGCCAGGCUGAACGGAACCAACAUUACUCAAUGUCAACAUCAAAAUUGCUUUGGAAU .(((((((((.(((........................((((......))))..........((((...))))..))).....(((((....)))))...........)))))))))... (-21.99 = -22.05 + 0.06)

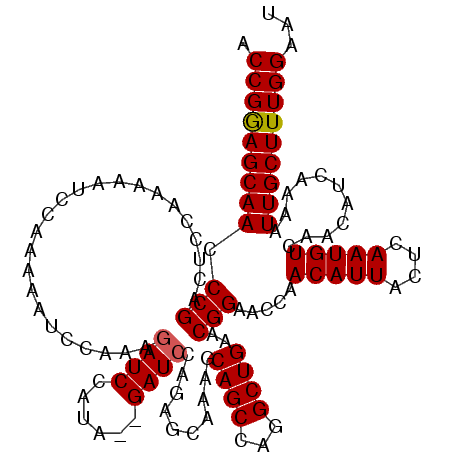
| Location | 15,629,400 – 15,629,518 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.84 |
| Mean single sequence MFE | -34.44 |
| Consensus MFE | -26.56 |
| Energy contribution | -26.50 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15629400 118 - 23771897 AUUCCAAAGCAAUUUUGAUGUUGACAUUGAGUAAUGUUGGUUCCGUUCAGCCUGGCUGCUUUACUCUGGAUC--UAUGGAUCUCUGGAUUUUUGGAUUUUUGGAAUCGGGUUGCUCCGGU ...((..((((((((..(((....)))..)).....((((((((((..(((......)))..))...(((((--((..((((....))))..)))))))..))))))))))))))..)). ( -30.90) >DroSec_CAF1 51628 118 - 1 AUUCCAAAGCAAUUUUGAUGUUGACAUUGAGUAAUGUUGGUUCCGUUCAGCCUGGCUGCUUUGCUCUGGAUC--UAUGGAUCUCUGGAUUUUGGGAUUUUUGAAGUCGGGUUGCUCCGGU ...((((......((..(((....)))..)).....))))..(((..((((((((((......(((..((((--((........))))))..)))........))))))))))...))). ( -33.14) >DroSim_CAF1 52637 118 - 1 AUUCCAAAGCAAUUUUGAUGUUGACAUUGAGUAAUGUUGGUUCCGUUCAGCCUGGCUGCUUUGCUCUGGAUC--UAUGGAUCUUAGGAUUUUUGGAUUUUUGGAGUCGGGUUGCUCCGGU ...((((......((..(((....)))..)).....))))..(((..((((((((((.(........(((((--((..((((....))))..)))))))..).))))))))))...))). ( -34.40) >DroYak_CAF1 56196 120 - 1 AUUCCAAAGCAAUUUUGAUGUUGACAUUGAGUAAUGUUGCUUCCGUUCAGCCUGGCUGCUUUACUCUGGAUCUCUAUGGAUUUUUAGAUCUGUGGAUUUUUGGAGUCGGGUUGCUUCGGU ......(((((((((..(((....)))..))....)))))))(((..(((((((........((((..((..((((((((((....))))))))))..))..)))))))))))...))). ( -39.30) >consensus AUUCCAAAGCAAUUUUGAUGUUGACAUUGAGUAAUGUUGGUUCCGUUCAGCCUGGCUGCUUUACUCUGGAUC__UAUGGAUCUCUGGAUUUUUGGAUUUUUGGAGUCGGGUUGCUCCGGU ...((((......((..(((....)))..)).....))))..(((..((((((((((.(....((..(((((......)))))..))(((....)))....).))))))))))...))). (-26.56 = -26.50 + -0.06)

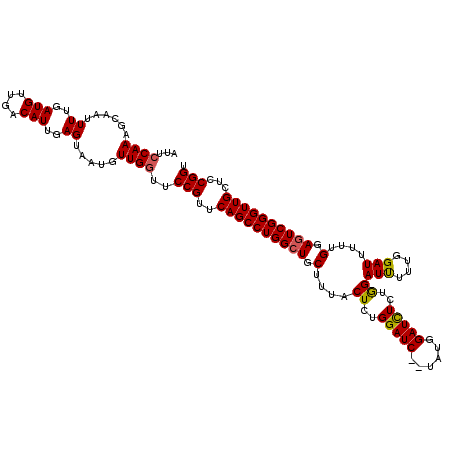
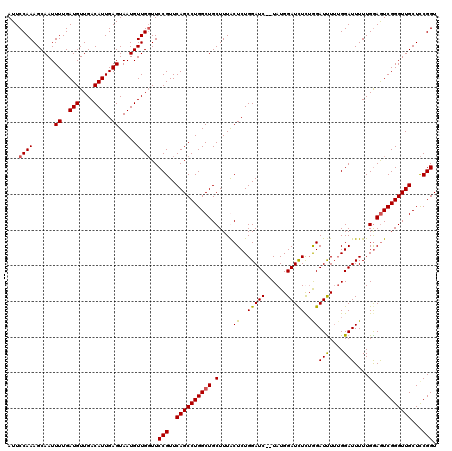
| Location | 15,629,440 – 15,629,558 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.18 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -24.15 |
| Energy contribution | -23.90 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15629440 118 + 23771897 UCCAUA--GAUCCAGAGUAAAGCAGCCAGGCUGAACGGAACCAACAUUACUCAAUGUCAACAUCAAAAUUGCUUUGGAAUUCAUGCAUUCAUUUAUUUCGCCUUCAACAAAAAGGCAAAC ..(((.--.(((((((((((..((((...))))..........(((((....)))))...........))))))))).))..)))..............(((((.......))))).... ( -23.50) >DroSec_CAF1 51668 118 + 1 UCCAUA--GAUCCAGAGCAAAGCAGCCAGGCUGAACGGAACCAACAUUACUCAAUGUCAACAUCAAAAUUGCUUUGGAAUUCAUGCAUUCAUUUAUUUCGCCUUCAACAAAAAGGCAAAC ..(((.--.(((((((((((..((((...))))..........(((((....)))))...........))))))))).))..)))..............(((((.......))))).... ( -25.50) >DroSim_CAF1 52677 118 + 1 UCCAUA--GAUCCAGAGCAAAGCAGCCAGGCUGAACGGAACCAACAUUACUCAAUGUCAACAUCAAAAUUGCUUUGGAAUUCAUGCAUUCAUUUAUUUCGCCUUCAACAAAAAGGCAAAC ..(((.--.(((((((((((..((((...))))..........(((((....)))))...........))))))))).))..)))..............(((((.......))))).... ( -25.50) >DroYak_CAF1 56236 120 + 1 UCCAUAGAGAUCCAGAGUAAAGCAGCCAGGCUGAACGGAAGCAACAUUACUCAAUGUCAACAUCAAAAUUGCUUUGGAAUUCAUGCAUUCAUUUAUUUCGCCUUCAACAAAAAGGCAAAC ..(((.(((.((((((((((.((..((.(......)))..)).(((((....)))))...........)))))))))).))))))..............(((((.......))))).... ( -26.40) >consensus UCCAUA__GAUCCAGAGCAAAGCAGCCAGGCUGAACGGAACCAACAUUACUCAAUGUCAACAUCAAAAUUGCUUUGGAAUUCAUGCAUUCAUUUAUUUCGCCUUCAACAAAAAGGCAAAC ..........((((((((((..((((...))))..........(((((....)))))...........)))))))))).....................(((((.......))))).... (-24.15 = -23.90 + -0.25)

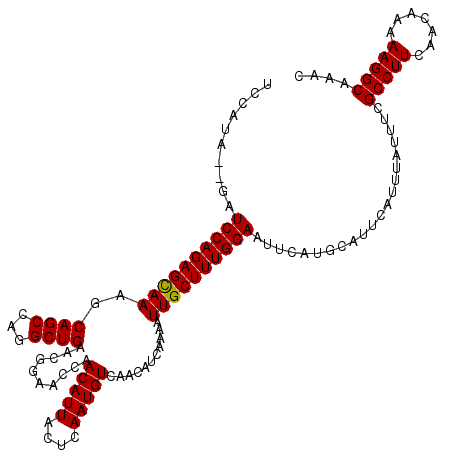
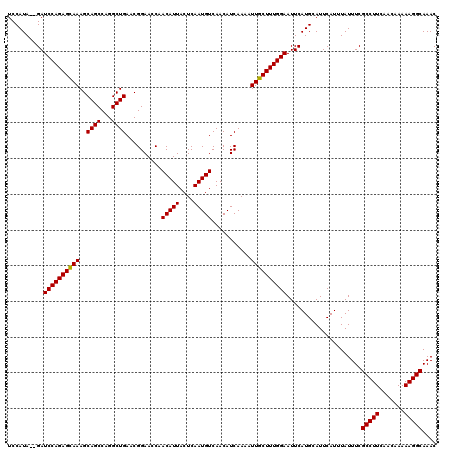
| Location | 15,629,478 – 15,629,598 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.58 |
| Mean single sequence MFE | -21.60 |
| Consensus MFE | -21.60 |
| Energy contribution | -21.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15629478 120 + 23771897 CCAACAUUACUCAAUGUCAACAUCAAAAUUGCUUUGGAAUUCAUGCAUUCAUUUAUUUCGCCUUCAACAAAAAGGCAAACUAACGCAGUAUAGAAAUUUUCAUUUUCGGCUGAGUUUUAA ........(((((..(((.........(((((...(((((..(((....)))..)))))(((((.......)))))........)))))...((((.......))))))))))))..... ( -21.60) >DroSec_CAF1 51706 120 + 1 CCAACAUUACUCAAUGUCAACAUCAAAAUUGCUUUGGAAUUCAUGCAUUCAUUUAUUUCGCCUUCAACAAAAAGGCAAACUAACGCAGUAUAGAAAUUUUCAUUUUCGGCUGAGUUUUAA ........(((((..(((.........(((((...(((((..(((....)))..)))))(((((.......)))))........)))))...((((.......))))))))))))..... ( -21.60) >DroSim_CAF1 52715 120 + 1 CCAACAUUACUCAAUGUCAACAUCAAAAUUGCUUUGGAAUUCAUGCAUUCAUUUAUUUCGCCUUCAACAAAAAGGCAAACUAACGCAGUAUAGAAAUUUUCAUUUUCGGCUGAGUUUUAA ........(((((..(((.........(((((...(((((..(((....)))..)))))(((((.......)))))........)))))...((((.......))))))))))))..... ( -21.60) >DroYak_CAF1 56276 120 + 1 GCAACAUUACUCAAUGUCAACAUCAAAAUUGCUUUGGAAUUCAUGCAUUCAUUUAUUUCGCCUUCAACAAAAAGGCAAACUAACGCAGUAUAGAAAUUUUCAUUUUCGGCUGAGUUUUAA ........(((((..(((.........(((((...(((((..(((....)))..)))))(((((.......)))))........)))))...((((.......))))))))))))..... ( -21.60) >consensus CCAACAUUACUCAAUGUCAACAUCAAAAUUGCUUUGGAAUUCAUGCAUUCAUUUAUUUCGCCUUCAACAAAAAGGCAAACUAACGCAGUAUAGAAAUUUUCAUUUUCGGCUGAGUUUUAA ........(((((..(((.........(((((...(((((..(((....)))..)))))(((((.......)))))........)))))...((((.......))))))))))))..... (-21.60 = -21.60 + -0.00)

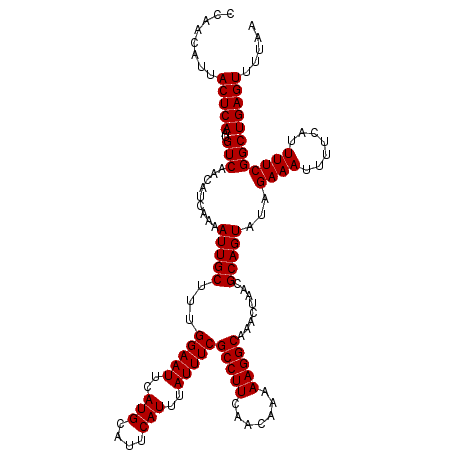
| Location | 15,629,558 – 15,629,672 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.31 |
| Mean single sequence MFE | -33.98 |
| Consensus MFE | -32.54 |
| Energy contribution | -32.60 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15629558 114 + 23771897 UAACGCAGUAUAGAAAUUUUCAUUUUCGGCUGAGUUUUAAUUCGGGGGAAUCGGAAUCUCCGGGGUCCUUAAAUAGCUGAACAGCCGUUUGGGAAUGAGAAUGGGAACGG------GAAC ...(.(.........(((((((((((((((((((((...(((..((((..((((.....))))..))))..)))))))...))))))....)))))))))))(....)).------)... ( -32.70) >DroSec_CAF1 51786 120 + 1 UAACGCAGUAUAGAAAUUUUCAUUUUCGGCUGAGUUUUAAUUCGGGGGAAUCGGAAUCUCCGGGGUCCUUAAAUAGCUGAACAGCCGGUUGGGAAUGGGAAUGGGAACGGGAAUGGGAAC ...(.((........(((..((((..((((((.......(((..((((..((((.....))))..))))..))).(((....)))))))))..))))..)))(....).....)).)... ( -35.10) >DroSim_CAF1 52795 114 + 1 UAACGCAGUAUAGAAAUUUUCAUUUUCGGCUGAGUUUUAAUUCGGGGGAAUCGGAAUCUCCGGGGUCCUUAAAUAGCUGAACAGCCGGUUGGGAAUGGGAAUGGGAACGG------GAAC ...(.(.........(((..((((..((((((.......(((..((((..((((.....))))..))))..))).(((....)))))))))..))))..)))(....)).------)... ( -34.40) >DroYak_CAF1 56356 104 + 1 UAACGCAGUAUAGAAAUUUUCAUUUUCGGCUGAGUUUUAAUUCGGGGGAAUCGGAAUCUCCGGGGUCCUUAAAUAGCUGAACAGCCGGUUGGGAAUGGGAAUGG---------------- ...............(((..((((..((((((.......(((..((((..((((.....))))..))))..))).(((....)))))))))..))))..)))..---------------- ( -33.70) >consensus UAACGCAGUAUAGAAAUUUUCAUUUUCGGCUGAGUUUUAAUUCGGGGGAAUCGGAAUCUCCGGGGUCCUUAAAUAGCUGAACAGCCGGUUGGGAAUGGGAAUGGGAACGG______GAAC ...............(((((((((..((((((.......(((..((((..((((.....))))..))))..))).(((....)))))))))..))))))))).................. (-32.54 = -32.60 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:50 2006