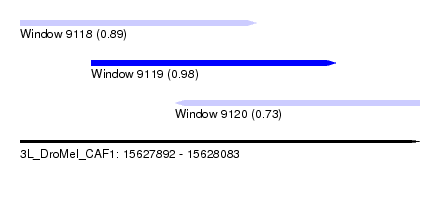
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,627,892 – 15,628,083 |
| Length | 191 |
| Max. P | 0.980522 |

| Location | 15,627,892 – 15,628,005 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 96.19 |
| Mean single sequence MFE | -33.85 |
| Consensus MFE | -29.06 |
| Energy contribution | -29.88 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15627892 113 + 23771897 AAAUAAAAAUAAAAAUCAAUUUCAGGGCAUUUCCUGGCCACGUUGCACGUGCCACGUGCCAAAACAACAACAAUAACA-ACUCGAGUGGAGUUUGGCUGGCUGGGGGGAUUCCC ........................(((..(..((..(((((((.((....)).))))(((((................-((((.....))))))))).)))..))..)...))) ( -31.59) >DroSec_CAF1 50139 113 + 1 AAAUAAAAAUAAAAAUCAAUUUCAGGGCAUUUCCUGGCCACGUUGCACGUGCCACGUGCCAAAACAACAACAAUAACAAACUCGAGUGGGGUUUGGCUGGCUGGG-GGAUUCCC ........................(((.((..((..((((.((.(((((.....))))).................(((((((.....)))))))))))))..))-..)).))) ( -37.50) >DroSim_CAF1 51151 114 + 1 AAAUAAAAAUAAAAAUCAAUUUCAGGGCAUUUCCUGGCCACGUUGCACGUGCCACGUGCCAAAACAACAACAAUAACAAACUCGAGUGGAGUUUGGCUGACUGGGGGGAUUCCC ........................(((.((..((((((.((((.((....)).)))))))................(((((((.....)))))))........)))..)).))) ( -32.10) >DroYak_CAF1 54719 112 + 1 AAAUAAAAAUAAAAAUCAAUUUCAGGGCAUUUCCUGGCCACGUUGCACGUGCCACGUGCCAAAGGAACAACAAUAACA-ACUCGAGUGGAGUUUCGUUGGCUGGG-GGAUUCCC ........................(((.((..((..((((((..(((((.....)))))..................(-((((.....))))).)).))))..))-..)).))) ( -34.20) >consensus AAAUAAAAAUAAAAAUCAAUUUCAGGGCAUUUCCUGGCCACGUUGCACGUGCCACGUGCCAAAACAACAACAAUAACA_ACUCGAGUGGAGUUUGGCUGGCUGGG_GGAUUCCC ........................(((.((((((..(((((...(((((.....))))).................(((((((.....)))))))).))))..)).)))).))) (-29.06 = -29.88 + 0.81)



| Location | 15,627,926 – 15,628,043 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -36.52 |
| Consensus MFE | -29.52 |
| Energy contribution | -29.24 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15627926 117 + 23771897 UGGCCACGUUGCACGUGCCACGUGCCAAAACAACAACAAUAACA-ACUCGAGUGGAGUUUGGCUGGCUGGGGGGAUUCCCCCAUGUACAUG--UAUAUGCUGAUAGCACUCGAACCCAAA ((((.((((.((....)).)))))))).................-..(((((((....(..((.....(((((....)))))(((((....--)))))))..)...)))))))....... ( -39.20) >DroSec_CAF1 50173 115 + 1 UGGCCACGUUGCACGUGCCACGUGCCAAAACAACAACAAUAACAAACUCGAGUGGGGUUUGGCUGGCUGGG-GGAUUCCCCCAUACACA----UAUAUGCUGAUAGCACUCGAACCCAAA ((((.((((.((....)).))))))))....................(((((((....(..((((..((((-((...))))))..))..----.....))..)...)))))))....... ( -36.71) >DroSim_CAF1 51185 116 + 1 UGGCCACGUUGCACGUGCCACGUGCCAAAACAACAACAAUAACAAACUCGAGUGGAGUUUGGCUGACUGGGGGGAUUCCCCCAUACACA----UAUAUGCUGAUAGCACUCGAACCCAAA ((((.((((.((....)).))))))))....................(((((((....(..((((..((((((....))))))..))..----.....))..)...)))))))....... ( -36.41) >DroEre_CAF1 51419 109 + 1 UGGCCACGUUGCACGUGCCACGUGCCAAAACAACAACAAUAACA-ACUCGAGUGGAGCUGCCU-CGCUGGG-GUAUUCCCCCAUA--------UAUAUGCUGAUAGCACUCGAACCCAAA ((((.((((.((....)).)))))))).................-..(((((((.(((.....-.)))(((-(.....))))...--------.............)))))))....... ( -32.70) >DroYak_CAF1 54753 118 + 1 UGGCCACGUUGCACGUGCCACGUGCCAAAGGAACAACAAUAACA-ACUCGAGUGGAGUUUCGUUGGCUGGG-GGAUUCCCCCAUAUACUCGUAUAUAUGCUGAUAGCACUCGAACCCAAA ((((.((((.((....)).)))))))).................-..((((((((.....)((..((.(((-((...)))))(((((....)))))..))..))..)))))))....... ( -37.60) >consensus UGGCCACGUUGCACGUGCCACGUGCCAAAACAACAACAAUAACA_ACUCGAGUGGAGUUUGGCUGGCUGGG_GGAUUCCCCCAUACACA____UAUAUGCUGAUAGCACUCGAACCCAAA ((((.((((.((....)).))))))))....................(((((((.((((.....))))(((.(.....))))........................)))))))....... (-29.52 = -29.24 + -0.28)



| Location | 15,627,966 – 15,628,083 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.57 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -22.42 |
| Energy contribution | -23.66 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15627966 117 - 23771897 AGAGGGCACAAAACUGCUACCCCCAAAAUGGUAAUGAAAUUUUGGGUUCGAGUGCUAUCAGCAUAUA--CAUGUACAUGGGGGAAUCCCCCCAGCCAGCCAAACUCCACUCGAGU-UGUU ....((((......))))...((((((((.........))))))))((((((((......((((...--.))))...((((((....)))))).............)))))))).-.... ( -34.70) >DroSec_CAF1 50213 115 - 1 AGAGGGCACAAAACUGCUACCCCCAAAAUGGUAAUGAAAUUUUGGGUUCGAGUGCUAUCAGCAUAUA----UGUGUAUGGGGGAAUCC-CCCAGCCAGCCAAACCCCACUCGAGUUUGUU ....((((......))))...((((((((.........))))))))((((((((......((((...----.)))).((((((...))-)))).............))))))))...... ( -33.60) >DroSim_CAF1 51225 116 - 1 AGAGGGCACAAAACUGCUACCCCCAAAAUGGUAAUGAAAUUUUGGGUUCGAGUGCUAUCAGCAUAUA----UGUGUAUGGGGGAAUCCCCCCAGUCAGCCAAACUCCACUCGAGUUUGUU .((..(((((....((((...((((((((.........))))))))...((......))))))....----))))).((((((....)))))).))...(((((((.....))))))).. ( -36.60) >DroEre_CAF1 51459 90 - 1 ------------------ACCCCGAAAA-GGUAAUCAAAUUUUGGGUUCGAGUGCUAUCAGCAUAUA--------UAUGGGGGAAUAC-CCCAGCG-AGGCAGCUCCACUCGAGU-UGUU ------------------..(((((((.-.(....)...)))))))((((.((((.....))))...--------..(((((.....)-)))).))-))(((((((.....))))-))). ( -29.00) >DroYak_CAF1 54793 117 - 1 AGAGGGCACAAAA-UGCUACCCCAAAAAUGGUAAUGAAAUUUUGGGUUCGAGUGCUAUCAGCAUAUAUACGAGUAUAUGGGGGAAUCC-CCCAGCCAACGAAACUCCACUCGAGU-UGUU .(..(((.....(-((((..(((((((.(........).))))))).....((((.....)))).......))))).((((((...))-)))))))..)..(((((.....))))-)... ( -32.60) >consensus AGAGGGCACAAAACUGCUACCCCCAAAAUGGUAAUGAAAUUUUGGGUUCGAGUGCUAUCAGCAUAUA____UGUAUAUGGGGGAAUCC_CCCAGCCAGCCAAACUCCACUCGAGU_UGUU ....((((......))))...((((((((.........))))))))((((((((.......((((((......))))))(((.......)))..............))))))))...... (-22.42 = -23.66 + 1.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:44 2006