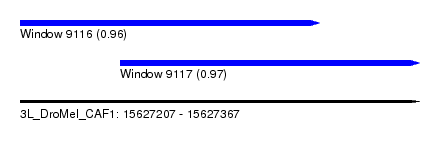
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,627,207 – 15,627,367 |
| Length | 160 |
| Max. P | 0.967031 |

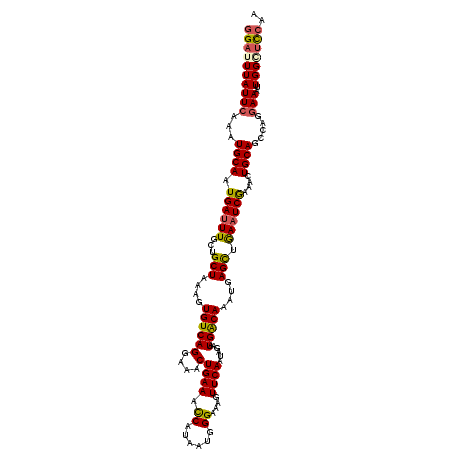
| Location | 15,627,207 – 15,627,327 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.17 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -24.46 |
| Energy contribution | -25.10 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15627207 120 + 23771897 UUGUCAGUGUUUUUGUAGCAAGUGAAGGCAACUAUUUUGGGGAUUUAUUCAAAUGCAAUGAUUUUCUGCUAAAGUGUCAGGAAACUGAAAUCAUAAUGGGAAAGUUCAAUAGAAUGACAA .(((((...((.(((..(((..((((..(..(......)..).....))))..)))...((((((((.........((((....))))..........))))))))))).))..))))). ( -25.11) >DroSec_CAF1 49448 120 + 1 UUAUCAGUGUUUUUAUUGCAAGUGAAGGCAACUAUUUUGGGGAUUUAUUCAAAUGCAAUGAUUUGCUGCUUAAAUGUCAGGAAACUGAAACCAUAAUUGGAAAGUUCAAUAGAAUGACAA ....(((((...((((((((..((((..(..(......)..).....))))..))))))))..)))))........((((....))))..(((....)))...((((....))))..... ( -26.90) >DroSim_CAF1 50451 120 + 1 UUUUCAGUGUUUUUAUAGCAAGUGAAGGCAACUAUUUUGGGGAUUUAUUCAAAUGCAGUGAUUUGCUGCUUAAAUGUCAGGAAACUGAAACCAUAAUGGGAAAGUUCAAUAGAAUGACAA .((((..(((((((((.....))))))))).(((((.(((..............(((((.....))))).......((((....))))..))).)))))))))((((....))))..... ( -29.90) >DroEre_CAF1 50691 120 + 1 UUGUCAGUGUUUCUACAGGAAGUGAAGGCAGCUAUUUUGGGGAUUUAUUGAAAUGCAAUGAUUUGUUGCUAAAGUGUCAGGAAACUGAAACCAUAAUGGGCAAGUUCAAUGGAAUGGCAA .((((..(..(((.....)))..)..))))((((((..(..((.((((((.....))))))....(((((...(((((((....))))...)))....)))))..))..)..)))))).. ( -30.40) >DroYak_CAF1 54003 120 + 1 UUGUCAGUGUUUUUAUAGGAAGUGAGUGCAACUAUUUUGGAGAUUUAUUCAAAUGCAAUGACUUGUUGCUAAAGUGUCAGGAAACUGAAACCAUAAUGGGGAAGUUCAAUAAAAUGACAA .(((((.(((.(((((.....))))).))).(((((.(((..............(((((.....))))).......((((....))))..))).)))))...............))))). ( -26.70) >consensus UUGUCAGUGUUUUUAUAGCAAGUGAAGGCAACUAUUUUGGGGAUUUAUUCAAAUGCAAUGAUUUGCUGCUAAAGUGUCAGGAAACUGAAACCAUAAUGGGAAAGUUCAAUAGAAUGACAA .(((((.(((((((((.....))))))))).(((((.(((..............(((((.....))))).......((((....))))..))).)))))...............))))). (-24.46 = -25.10 + 0.64)

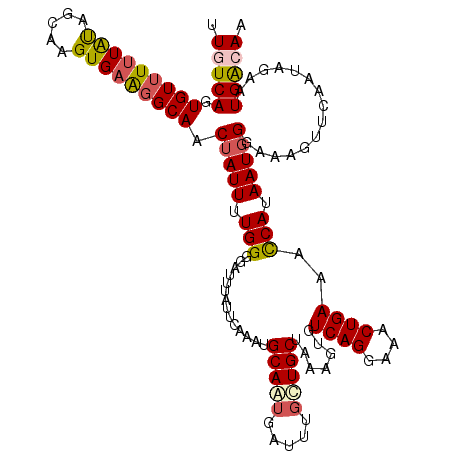
| Location | 15,627,247 – 15,627,367 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.75 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -22.64 |
| Energy contribution | -22.64 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.967031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15627247 120 + 23771897 GGAUUUAUUCAAAUGCAAUGAUUUUCUGCUAAAGUGUCAGGAAACUGAAAUCAUAAUGGGAAAGUUCAAUAGAAUGACAAAUGAGUUGAAUCGAAACUGCAGCCAGGAAAUUGGGUCCAA ((((((((((...((((....((((((..((..((.((((....)))).))..))..))))))(((((((...((.....))..)))))))......)))).....)))..))))))).. ( -26.50) >DroSec_CAF1 49488 120 + 1 GGAUUUAUUCAAAUGCAAUGAUUUGCUGCUUAAAUGUCAGGAAACUGAAACCAUAAUUGGAAAGUUCAAUAGAAUGACAAAUGAGCUAAAUCGAAACUGCAGCCAGGAAAUUGGAUCCAA ((((((((((...((((.(((((((..(((((..((((((....)((((.(((....)))....))))......)))))..))))))))))))....)))).....)))..))))))).. ( -31.10) >DroSim_CAF1 50491 120 + 1 GGAUUUAUUCAAAUGCAGUGAUUUGCUGCUUAAAUGUCAGGAAACUGAAACCAUAAUGGGAAAGUUCAAUAGAAUGACAAAUGAGCUAAAUCAUAACUGCAGCCAGGAAAUUGGGUUCAA ((((((((((...((((((((((((..(((((..((((((....)((((.((.....)).....))))......)))))..)))))))))))...)))))).....)))..))))))).. ( -30.60) >DroEre_CAF1 50731 120 + 1 GGAUUUAUUGAAAUGCAAUGAUUUGUUGCUAAAGUGUCAGGAAACUGAAACCAUAAUGGGCAAGUUCAAUGGAAUGGCAAAUGAGCUGAAUCAAAACUGCAGUCAGGAAAUUGGCUCCAA (((..........((((.((((((..((((...(((((((....))))...)))....))))((((((.((......))..))))))))))))....))))(((((....)))))))).. ( -29.20) >DroYak_CAF1 54043 120 + 1 AGAUUUAUUCAAAUGCAAUGACUUGUUGCUAAAGUGUCAGGAAACUGAAACCAUAAUGGGGAAGUUCAAUAAAAUGACAAAUGAGCUGAAUCGAAACUGCAGUCGGGAAAUUGGCCCCAA ..............(((((.....)))))....((.((((....)))).)).....(((((.((((((.............))))))...((((........))))........))))). ( -26.62) >consensus GGAUUUAUUCAAAUGCAAUGAUUUGCUGCUAAAGUGUCAGGAAACUGAAACCAUAAUGGGAAAGUUCAAUAGAAUGACAAAUGAGCUGAAUCGAAACUGCAGCCAGGAAAUUGGCUCCAA ((((((((((...((((.((((((...(((....((((((....)((((.((......))....))))......)))))....))).))))))....)))).....)))..))))))).. (-22.64 = -22.64 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:41 2006