
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,621,551 – 15,621,737 |
| Length | 186 |
| Max. P | 0.804904 |

| Location | 15,621,551 – 15,621,666 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.94 |
| Mean single sequence MFE | -24.27 |
| Consensus MFE | -20.50 |
| Energy contribution | -20.34 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
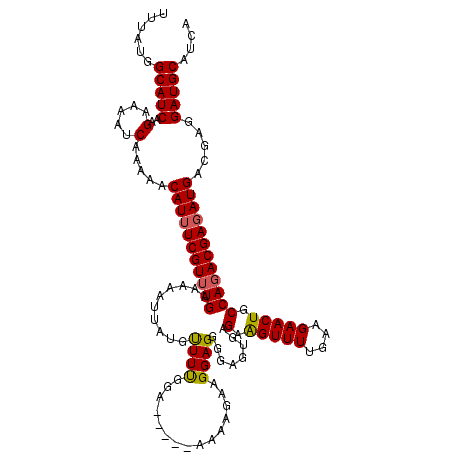
>3L_DroMel_CAF1 15621551 115 + 23771897 UUUAUGGCAUCAAGAAAAUCAAAAACAUUUCGUUUGAAAAAUUAUGUUUUGGA-----GUAAGAAGGAGUGGAGUGAGAAGUUUUGAGGAACUGCCAGACGAGAUGACGAGGAUGCAUCA ......(((((..(.....).....(((((((((((........(.((((...-----...)))).)............(((((....)))))..))))))))))).....))))).... ( -22.50) >DroSec_CAF1 44350 115 + 1 UUUAUGGCAUCAAGAAAAUCAAAAACAUUUCGUUUGAAAAAUUAUGCUUUGGA-----AAAAGAUGGAGGGGAGUGAGGAGUUUUGAAGAACUGCCAGACGAGAUGACGAGGAUGCAUCA ......(((((..(.....).....(((((((((((.....((((.((((...-----.))))))))..........(.(((((....))))).)))))))))))).....))))).... ( -27.20) >DroSim_CAF1 44921 115 + 1 UUUAUGGCAUCAAGAAAAUCAAAAACAUUUCGUUUGAAAAAUUAUGCUUUGGA-----AAAAGAUGGAGGGGAGUGAGAAGUUUUGAAGAACUGCCAGACGAGAUGACGAGGAUGCAUCA ......(((((..(.....).....(((((((((((.....((((.((((...-----.))))))))............(((((....)))))..))))))))))).....))))).... ( -25.70) >DroEre_CAF1 45007 115 + 1 UUUAUGGCAUCAAGAAAAUCAAAAACAUUUCGUUUGAAAAAUUAUGUUUCGGG-----AUGAAAAGGAGGGCAGGGAGAGGUUUUGAAGAACUGCCAGACGAUAUGACGAGGAUGCAUCA ......(((((..(.....).....(((.((((((((((.......))))...-----...........(((((.................))))))))))).))).....))))).... ( -22.53) >DroYak_CAF1 47558 119 + 1 UUUAUGGCAUCAAGAAAAUCAAAAACAUUUCGU-UGAAAAAUUAUGUUUUGGAAAGCGAAGGAGAGGAGGGGAGGGAGGAGUUUUGAAGAACUGCCAGACGAGAUGACGAGGAUGCAUCA ......(((((..(.....).....((((((((-(.........(.(((((.....))))).)..............(((((((....))))).)).))))))))).....))))).... ( -23.40) >consensus UUUAUGGCAUCAAGAAAAUCAAAAACAUUUCGUUUGAAAAAUUAUGUUUUGGA_____AAAAGAAGGAGGGGAGUGAGAAGUUUUGAAGAACUGCCAGACGAGAUGACGAGGAUGCAUCA ......(((((..(.....).....(((((((((((..........((((...............))))........(.(((((....))))).)))))))))))).....))))).... (-20.50 = -20.34 + -0.16)


| Location | 15,621,626 – 15,621,737 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.24 |
| Mean single sequence MFE | -31.96 |
| Consensus MFE | -30.68 |
| Energy contribution | -30.72 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15621626 111 - 23771897 GUCCAGACCAGCAUCGCGAUGAGUUGAGCCAGGCACGCACA---------UCGUGCACAUCGUCAUGCUUGUCUUACACUUGAUGCAUCCUCGUCAUCUCGUCUGGCAGUUCCUCAAAAC (.((((((.(((((.((((((.(.....)...(((((....---------.))))).)))))).)))))...........(((((......)))))....)))))))............. ( -31.30) >DroSec_CAF1 44425 111 - 1 GUCCAGACCAGCAUCGCGAUGAGUUGAGCCAGGCACGCACA---------UCGUGCACAUCGUCAUGCUUGUCUUACACUUGAUGCAUCCUCGUCAUCUCGUCUGGCAGUUCUUCAAAAC (.((((((.(((((.((((((.(.....)...(((((....---------.))))).)))))).)))))...........(((((......)))))....)))))))............. ( -31.30) >DroSim_CAF1 44996 111 - 1 GUCCAGACCAGCAUCGCGAUGAGUUGAGCCAGGCACGCACA---------UCGUGCACAUCGUCAUGCUUGUCUUGCACUUGAUGCAUCCUCGUCAUCUCGUCUGGCAGUUCUUCAAAAC (.((((((.(((((.((((((.(.....)...(((((....---------.))))).)))))).)))))...........(((((......)))))....)))))))............. ( -31.30) >DroEre_CAF1 45082 120 - 1 CUCCAGACCAGCAUCGCGAUGAGUUGAGCCAGGCACGCACAUCGUCCACAUCGUGCACAUCGUCAUGCUUGUCUUACACUUGAUGCAUCCUCGUCAUAUCGUCUGGCAGUUCUUCAAAAC ((((((((.(((((.((((((.(((......)))..((((............)))).)))))).)))))...........(((((......)))))....)))))).))........... ( -31.80) >DroYak_CAF1 47637 111 - 1 GUCCAGACCAGCAUCGCGAUGAGCUGAGCCCGGCACGCACA---------UCGGGCACAUCGUCAUGCUUGUCUUGCACUUGAUGCAUCCUCGUCAUCUCGUCUGGCAGUUCUUCAAAAC (.((((((..((...))((((((.((.((((((........---------)))))).(((((...(((.......)))..)))))))..)))))).....)))))))............. ( -34.10) >consensus GUCCAGACCAGCAUCGCGAUGAGUUGAGCCAGGCACGCACA_________UCGUGCACAUCGUCAUGCUUGUCUUACACUUGAUGCAUCCUCGUCAUCUCGUCUGGCAGUUCUUCAAAAC ..((((((.(((((.((((((.(((......)))..((((............)))).)))))).)))))...........(((((......)))))....)))))).............. (-30.68 = -30.72 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:39 2006