
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,619,955 – 15,620,115 |
| Length | 160 |
| Max. P | 0.824415 |

| Location | 15,619,955 – 15,620,075 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.08 |
| Mean single sequence MFE | -48.12 |
| Consensus MFE | -31.44 |
| Energy contribution | -32.60 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
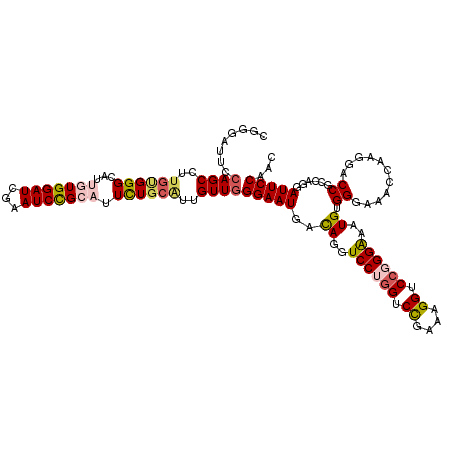
>3L_DroMel_CAF1 15619955 120 + 23771897 CGGGAUUCCAGCCUUGUGGGCAUUGUGGAUCGAAUCCGCAUUCUGCAUUGUUGGGAAUGACAGGUCCUGGUCUGCAAGGUCCGGGAAAUGUGGGAGGCCAAGGACCGCCGGGAUUCCAAC .((((((((.((..((..((...(((((((...))))))).))..))((.((((...(.(((..((((((.((....)).))))))..))).)....)))).))..)).))))))))... ( -47.20) >DroSec_CAF1 42855 120 + 1 CGGGAUUCCAGCCUUGUGGGCAUUGUGGAUCGAAUCCGCAUUCUGCAUUGUUGGGAAUGACAGGUCCUGGUCCGGAAGGUCCGGGAAAUGGGGAAAAACAAGGACCGCCAGGAUUCCAAC .(((((((((((..((..((...(((((((...))))))).))..))..))))((.....((..((((((.((....)).))))))..))((............)).)).)))))))... ( -47.50) >DroSim_CAF1 43209 120 + 1 CGGGAUUCCAGCCUUGUGGGCAUUGUGGAUCGAAUCCGCAUUCUGCAUUGUUGGGAAUAACAGGUCCUGGUCCGGAAGGUCCGGGAAAUGGGGGAAAACACGGACCGCCAGGAUUCCAAC .(((((((((((..((..((...(((((((...))))))).))..))..)))))..........((((((.((....)).))))))..(((.((..........)).))).))))))... ( -47.80) >DroEre_CAF1 43374 120 + 1 CCGCACCGCAGCCUGGCGGGCAAUUUGGAUCAAAUCUGUUGUUUGGAUUGUUGGGAAUGACAGGUCCGGGGCCGAAAGGUCCGGGGUAUGUGGGUAUCCUAGGACCGUCAGGAUUCCAAC ..((.((((......))))))((((..((.(((.....)))))..))))(((((((.((((.(((((.(((((....)))))(((((((....))))))).)))))))))...))))))) ( -54.30) >DroYak_CAF1 46095 120 + 1 CGGCACUCCAGCCUUGCGGGCAGUCUGGAUCAAAUCCGUUUUCUCCGUUGUUCGGAAUGAUAGGUCCGAGACCGAAAGGUCCGGGAUUUGUGGGUAUCCAAGGACCGUCGAGAUUCCAAC ((((..(((.(((.....)))....(((((...((((......((((.....))))...((((((((..((((....))))..))))))))))))))))).)))..)))).......... ( -43.80) >consensus CGGGAUUCCAGCCUUGUGGGCAUUGUGGAUCGAAUCCGCAUUCUGCAUUGUUGGGAAUGACAGGUCCUGGUCCGAAAGGUCCGGGAAAUGUGGGAAACCAAGGACCGCCAGGAUUCCAAC ........((((..((((((...(((((((...))))))).))))))..))))(((((..((..((((((.((....)).))))))..)).((...........))......)))))... (-31.44 = -32.60 + 1.16)


| Location | 15,619,995 – 15,620,115 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.67 |
| Mean single sequence MFE | -47.92 |
| Consensus MFE | -35.94 |
| Energy contribution | -35.82 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.727816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
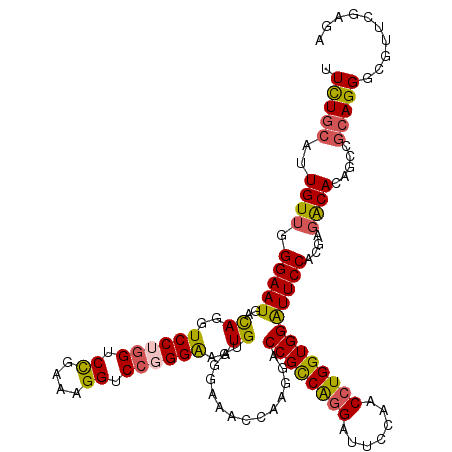
>3L_DroMel_CAF1 15619995 120 + 23771897 UUCUGCAUUGUUGGGAAUGACAGGUCCUGGUCUGCAAGGUCCGGGAAAUGUGGGAGGCCAAGGACCGCCGGGAUUCCAACCUGGUGGAUUCCACGAGACACACCCGCAGGGCGUUCGAGA .(((((....((((...(.(((..((((((.((....)).))))))..))).)....))))(((((((((((.......))))))))..))).............))))).......... ( -43.20) >DroSec_CAF1 42895 120 + 1 UUCUGCAUUGUUGGGAAUGACAGGUCCUGGUCCGGAAGGUCCGGGAAAUGGGGAAAAACAAGGACCGCCAGGAUUCCAACCUGGUGGAUUCCACGAGACACAGCCGCAGGGCGUUCGAGA ((((.(((((((......)))...((((((.((....)).)))))))))).))))......(((((((((((.......))))))))..))).((((.....(((....))).))))... ( -48.50) >DroSim_CAF1 43249 120 + 1 UUCUGCAUUGUUGGGAAUAACAGGUCCUGGUCCGGAAGGUCCGGGAAAUGGGGGAAAACACGGACCGCCAGGAUUCCAACCUGGUGGAUUCCACGAGACACAGCCGCAGGGCGUUCGAGA ((((.((((((((....))))...((((((.((....)).)))))))))).))))......(((((((((((.......))))))))..))).((((.....(((....))).))))... ( -48.70) >DroEre_CAF1 43414 120 + 1 GUUUGGAUUGUUGGGAAUGACAGGUCCGGGGCCGAAAGGUCCGGGGUAUGUGGGUAUCCUAGGACCGUCAGGAUUCCAACCUGGUGGGUUCCACGCGGCACAUCCGCAGGGCCUUCGAGG .........(((((((.((((.(((((.(((((....)))))(((((((....))))))).)))))))))...)))))))((.(.((((((...((((.....)))).)))))).).)). ( -55.80) >DroYak_CAF1 46135 120 + 1 UUCUCCGUUGUUCGGAAUGAUAGGUCCGAGACCGAAAGGUCCGGGAUUUGUGGGUAUCCAAGGACCGUCGAGAUUCCAACCUGGUGGAUUCCACGAUGCACAUCCGCAGGGCUUUCGAGG ...((((.....))))......((((((.((((....))))).((((........))))..))))).((((((..((......((((...))))..(((......)))))..)))))).. ( -43.40) >consensus UUCUGCAUUGUUGGGAAUGACAGGUCCUGGUCCGAAAGGUCCGGGAAAUGUGGGAAACCAAGGACCGCCAGGAUUCCAACCUGGUGGAUUCCACGAGACACAGCCGCAGGGCGUUCGAGA .(((((..((((.(((((..((..((((((.((....)).))))))..))..............((((((((.......)))))))))))))....)))).....))))).......... (-35.94 = -35.82 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:36 2006