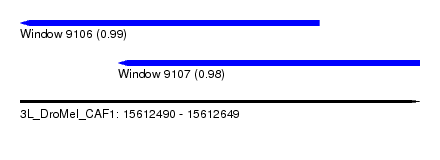
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,612,490 – 15,612,649 |
| Length | 159 |
| Max. P | 0.994716 |

| Location | 15,612,490 – 15,612,609 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.38 |
| Mean single sequence MFE | -45.86 |
| Consensus MFE | -44.46 |
| Energy contribution | -43.50 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
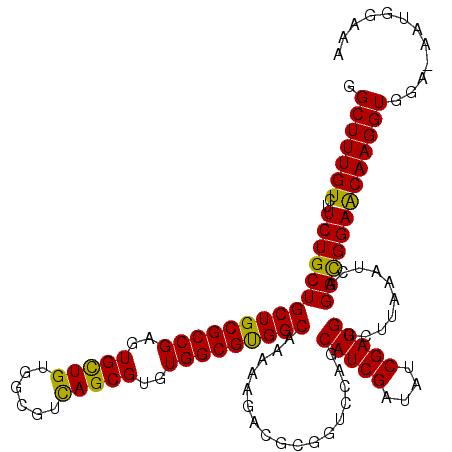
>3L_DroMel_CAF1 15612490 119 - 23771897 GGCUUUGUCUUCUGCUGCUGCGCCGAGUGCUGUGGCGUCAGCGUGUGGCGUGGCAAAAAGACACGGUCCAGCAUCGAUAUCGAUGGCUUAAAUCAGGCGGAACAAGGUGGA-AAUGGAAA .(((((((..((((((((..(((((..(((((......)))))..)))))..))..........(((..(((((((....)))).)))...))).)))))))))))))...-........ ( -46.10) >DroSec_CAF1 35569 120 - 1 GGCUUUGUCUUCUGCUGCUGCGCCGAGUGCUGUGGCGUCAGCGUGUGGCGUGGCAAAAAGACGCGGUCCAGCAUCGAUAUCGAUGGCUUAAAUCAGGCGGAGCAAGGUGGAAAAUGGAAA ...((..(.(((..((((..(((((..(((((......)))))..)))))..))..........(.(((..(((((....)))))((((.....))))))).)..))..))).)..)).. ( -47.00) >DroSim_CAF1 35613 119 - 1 GGCUUUGUCUUCUGCUGCUGCGCCGAGUGCUGUGGCGUCAGCGUGUGGCGUGGCAAAAAGACGCGGUCCAGCAUCGAUAUCGAUGGCUUAAAUCAGGCGGAACAAGGUGGA-AGUGGAAA .(((((((..((((((((..(((((..(((((......)))))..)))))..))..........(((..(((((((....)))).)))...))).)))))))))))))...-........ ( -46.10) >DroEre_CAF1 35675 119 - 1 GGCUUUGUCUUCUGCUGCUGCGCCGAGUGUUGUGGCGUUAGCGUGUGGCGCGGCAAGAAGUCGCUGUCCAGCAUCGAUAUCGAUGGCUUAAAUCAGGUGGAACAAGGUGGG-AAUGGCAA .(((((((..((..(((((((((((..(((((......)))))..)))))))))...((((((((....)))((((....)))))))))......))..)))))))))...-........ ( -45.50) >DroYak_CAF1 38069 119 - 1 GGCUUUGUCUUCUGCUGCUGCGCCGAGUGCUGCGGUGUCAGCGUGUGGCGCGGCCGAAAGUCACGGUCCAGCAUCGAUAUCGAUGGCCUUAAUCAGGUGGAACAAGGUGGA-AAUGGGAG .(((((((..((..(((((((((((..(((((......)))))..)))))))))(....)....(((....(((((....)))))))).......))..)))))))))...-........ ( -44.60) >consensus GGCUUUGUCUUCUGCUGCUGCGCCGAGUGCUGUGGCGUCAGCGUGUGGCGUGGCAAAAAGACGCGGUCCAGCAUCGAUAUCGAUGGCUUAAAUCAGGCGGAACAAGGUGGA_AAUGGAAA .(((((((..(((((((((((((((..(((((......)))))..))))))))).................(((((....)))))..........)))))))))))))............ (-44.46 = -43.50 + -0.96)


| Location | 15,612,529 – 15,612,649 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -47.32 |
| Consensus MFE | -44.78 |
| Energy contribution | -44.78 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15612529 120 - 23771897 CAUAGUCCUCAUGGUUUCCGCCUGGAUCCUGCUCAUUCUGGGCUUUGUCUUCUGCUGCUGCGCCGAGUGCUGUGGCGUCAGCGUGUGGCGUGGCAAAAAGACACGGUCCAGCAUCGAUAU ((..((((....(((....))).))))..)).((((.(((((((.((((((....(((..(((((..(((((......)))))..)))))..)))..)))))).))))))).)).))... ( -48.70) >DroSec_CAF1 35609 120 - 1 CAUAGUCCUCAUGGUCUCCGCCUGGAUCCUGCUCAUUAUGGGCUUUGUCUUCUGCUGCUGCGCCGAGUGCUGUGGCGUCAGCGUGUGGCGUGGCAAAAAGACGCGGUCCAGCAUCGAUAU ...((.((....)).))..((.((((((..((((.....))))...(((((....(((..(((((..(((((......)))))..)))))..)))..)))))..))))))))........ ( -47.10) >DroSim_CAF1 35652 120 - 1 CAUAGUCCUCAUGGUCUCCGCCUGGAUCCUGCUCAUUCUGGGCUUUGUCUUCUGCUGCUGCGCCGAGUGCUGUGGCGUCAGCGUGUGGCGUGGCAAAAAGACGCGGUCCAGCAUCGAUAU ((..((((....(((....))).))))..)).((((.(((((((.((((((....(((..(((((..(((((......)))))..)))))..)))..)))))).))))))).)).))... ( -48.00) >DroEre_CAF1 35714 120 - 1 CAUAGUCCUGAUGGUCUCCGCCUGGAUCCUGCUCAUUCUGGGCUUUGUCUUCUGCUGCUGCGCCGAGUGUUGUGGCGUUAGCGUGUGGCGCGGCAAGAAGUCGCUGUCCAGCAUCGAUAU ((..((((....(((....))).))))..)).((((.((((((..((.(((((..((((((((((..(((((......)))))..))))))))))))))).))..)))))).)).))... ( -46.50) >DroYak_CAF1 38108 120 - 1 CAUUGUCCUGAUGGUCUCCGCCUGGAUCCUGCUCAUCCUGGGCUUUGUCUUCUGCUGCUGCGCCGAGUGCUGCGGUGUCAGCGUGUGGCGCGGCCGAAAGUCACGGUCCAGCAUCGAUAU ...((((..(((((..(((....)))..)........(((((((.((.(((...(.(((((((((..(((((......)))))..))))))))).).))).)).))))))))))))))). ( -46.30) >consensus CAUAGUCCUCAUGGUCUCCGCCUGGAUCCUGCUCAUUCUGGGCUUUGUCUUCUGCUGCUGCGCCGAGUGCUGUGGCGUCAGCGUGUGGCGUGGCAAAAAGACGCGGUCCAGCAUCGAUAU ((..((((....(((....))).))))..)).((((.((((((..((((((....((((((((((..(((((......)))))..))))))))))..))))))..)))))).)).))... (-44.78 = -44.78 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:30 2006