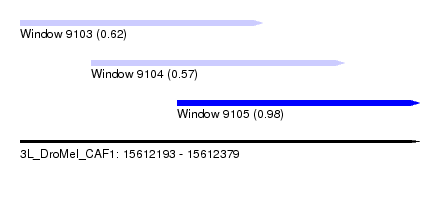
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,612,193 – 15,612,379 |
| Length | 186 |
| Max. P | 0.975880 |

| Location | 15,612,193 – 15,612,306 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.69 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -20.62 |
| Energy contribution | -20.26 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15612193 113 + 23771897 GAAU-------UUUAUAUAUAUCCAUUAAGCUCUGUGCUUAACCGAAUCGUAGAGUAUAUUGCACACUUGCAAAUUCUUGGAUAUCCCUGUGGCUGCUCGUUUAUUGUAGCCAUUUAUGG ....-------.......(((((((((((((.....)))))).........(((((...(((((....))))))))))))))))).((.((((((((.........))))))))....)) ( -29.70) >DroSec_CAF1 35264 113 + 1 GAAU-------UUUAUACAAAUCCUUUAGGCUCUGUGCUUAAGCGAAUCGGAGGGUAUAUUGCACACUUGCAAAUUCUUGGAUAUCCCUGUGGCUGCUCGUUUAUUGUAGCCAUUUAUGG ....-------..........((.(((((((.....))))))).))(((.((((((...(((((....))))))))))).)))...((.((((((((.........))))))))....)) ( -30.30) >DroSim_CAF1 35301 120 + 1 GAAUAUUUAUAUUUAUACACAUCCAUUAGGCUCUGUGCUUAAGCGAAUCGCAGGGUAUAUUGCACACUUGCAAAUUCUUGGAUAUCCCUGUGGCUGCUCGUUUAUUGUAGCCAUUUAUAG ....................(((((....((((((((.((.....)).))))))))...(((((....))))).....)))))......((((((((.........))))))))...... ( -29.60) >DroEre_CAF1 35380 105 + 1 --------------AUAUUUAUUUAUUGGGCU-UGUGCCUAGCAGAAUCGCAGGGUAUUUGGCGCACUUGAUAAAUCUUGGAUAUGCCUGUGGCUGCUCGCUUAUUGUAGCCAUUUAUGG --------------..(((((((...((.(((-.((((((.((......)).))))))..))).))...)))))))..........((.((((((((.........))))))))....)) ( -31.30) >DroYak_CAF1 37764 113 + 1 GAAU-------UUUAUAUUUAUUCAUUCAGCUUUGUGCUAAACUGAAUCGCAGGGUAUAUCGCACACUUGCUGGAUUCUGGAUAUCCCUGCGGCUGCUCGUUUAUUGUACCCAUUUAUGG ....-------.........(((((((.(((.....))).)).))))).((((((.((((((((....))).(....)..)))))))))))((.(((.........))).))........ ( -24.20) >consensus GAAU_______UUUAUAUAUAUCCAUUAGGCUCUGUGCUUAACCGAAUCGCAGGGUAUAUUGCACACUUGCAAAUUCUUGGAUAUCCCUGUGGCUGCUCGUUUAUUGUAGCCAUUUAUGG .....................((..((((((.....))))))..))...(((((((((.(((((....)))))........))).))))))((((((.........))))))........ (-20.62 = -20.26 + -0.36)


| Location | 15,612,226 – 15,612,344 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.65 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -24.02 |
| Energy contribution | -24.86 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15612226 118 + 23771897 AACCGAAUCGUAGAGUAUAUUGCACACUUGCAAAUUCUUGGAUAUCCCUGUGGCUGCUCGUUUAUUGUAGCCAUUUAUGGGA--UACUCGUGAAAUAUGAGUUUAGUUAUCCACAGGCAA ..((.......(((((...(((((....))))))))))((((((((((.((((((((.........))))))))....))))--.(((((((...))))))).....))))))..))... ( -35.10) >DroSec_CAF1 35297 118 + 1 AAGCGAAUCGGAGGGUAUAUUGCACACUUGCAAAUUCUUGGAUAUCCCUGUGGCUGCUCGUUUAUUGUAGCCAUUUAUGGGA--UACUCGUGAAAUAUGAGUUUAGUUAUCCACCGGCAA .......((((.(((((..(((((....))))).(((.((..((((((.((((((((.........))))))))....))))--))..)).))).............))))).))))... ( -39.30) >DroSim_CAF1 35341 120 + 1 AAGCGAAUCGCAGGGUAUAUUGCACACUUGCAAAUUCUUGGAUAUCCCUGUGGCUGCUCGUUUAUUGUAGCCAUUUAUAGGAUAUACUCGUGAAAUAUGAGUUUAGUUAUCCACCGGCAA ..((.....((((((....(((((....)))))(((....)))..))))))((((((.........)))))).......(((((.(((((((...))))))).....)))))....)).. ( -34.50) >DroEre_CAF1 35405 118 + 1 AGCAGAAUCGCAGGGUAUUUGGCGCACUUGAUAAAUCUUGGAUAUGCCUGUGGCUGCUCGCUUAUUGUAGCCAUUUAUGGGA--AACUCGUGAAAUAUGACCUUAGUUAUCCACAGGCAA ......(((.(((((((((.((....)).)))..))))))))).(((((((((((((.........)))))).(((((((..--..).))))))..(((((....)))))..))))))). ( -33.30) >DroYak_CAF1 37797 118 + 1 AACUGAAUCGCAGGGUAUAUCGCACACUUGCUGGAUUCUGGAUAUCCCUGCGGCUGCUCGUUUAUUGUACCCAUUUAUGGGA--UACUCGUGAAAUAUGACUUUAGUUAUCCACAGGCAA .......((((((((.((((((((....))).(....)..))))))))))))).((((........(((((((....)))).--)))..(((....(((((....))))).))).)))). ( -32.20) >consensus AACCGAAUCGCAGGGUAUAUUGCACACUUGCAAAUUCUUGGAUAUCCCUGUGGCUGCUCGUUUAUUGUAGCCAUUUAUGGGA__UACUCGUGAAAUAUGAGUUUAGUUAUCCACAGGCAA .........((.(((....(((((....)))))...)))(((((((((.((((((((.........))))))))....))))...(((((((...))))))).....)))))....)).. (-24.02 = -24.86 + 0.84)

| Location | 15,612,266 – 15,612,379 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.35 |
| Mean single sequence MFE | -36.86 |
| Consensus MFE | -29.50 |
| Energy contribution | -29.94 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15612266 113 + 23771897 GAUAUCCCUGUGGCUGCUCGUUUAUUGUAGCCAUUUAUGGGA--UACUCGUGAAAUAUGAGUUUAGUUAUCCACAGGCAAGGGAGA---ACUUGGCCAGUCUCC--CCAGGCCAAUCAAU ((((((((.((((((((.........))))))))....))))--)(((((((...))))))).............(((..((((((---.((.....)))))))--)...))).)))... ( -40.00) >DroSec_CAF1 35337 118 + 1 GAUAUCCCUGUGGCUGCUCGUUUAUUGUAGCCAUUUAUGGGA--UACUCGUGAAAUAUGAGUUUAGUUAUCCACCGGCAAGGGAGAACAACUUGGCCAGUCUCCCCCCAGGCCAAUCAAU ((((((((.((((((((.........))))))))....))))--)(((((((...))))))).............(((..((((((.(.....).....)))))).....))).)))... ( -37.40) >DroSim_CAF1 35381 120 + 1 GAUAUCCCUGUGGCUGCUCGUUUAUUGUAGCCAUUUAUAGGAUAUACUCGUGAAAUAUGAGUUUAGUUAUCCACCGGCAAGGGAGAACAACUUGGCCAGUCUCCCCCCAGGCCAAUCAAU (((.(((((((((((((.........)))))))).....(((((.(((((((...))))))).....))))).......)))))........(((((............))))))))... ( -37.60) >DroEre_CAF1 35445 113 + 1 GAUAUGCCUGUGGCUGCUCGCUUAUUGUAGCCAUUUAUGGGA--AACUCGUGAAAUAUGACCUUAGUUAUCCACAGGCAAGGGAGA---ACUUGGCCAGUCUCC--CCAGGCCAAUCAAU (((.(((((((((((((.........)))))).(((((((..--..).))))))..(((((....)))))..))))))).((((((---.((.....)))))))--).......)))... ( -38.20) >DroYak_CAF1 37837 113 + 1 GAUAUCCCUGCGGCUGCUCGUUUAUUGUACCCAUUUAUGGGA--UACUCGUGAAAUAUGACUUUAGUUAUCCACAGGCAAGGGAGA---ACUUGGCCAGUCUCC--CCAGGCCAAUCAAU (((.(((((...(((...........(((((((....)))).--)))..(((....(((((....))))).))).))).)))))..---...(((((.......--...))))))))... ( -31.10) >consensus GAUAUCCCUGUGGCUGCUCGUUUAUUGUAGCCAUUUAUGGGA__UACUCGUGAAAUAUGAGUUUAGUUAUCCACAGGCAAGGGAGA___ACUUGGCCAGUCUCC__CCAGGCCAAUCAAU (((.(((((((((((((.........)))))).((((((((.....))))))))......................)).)))))........(((((............))))))))... (-29.50 = -29.94 + 0.44)

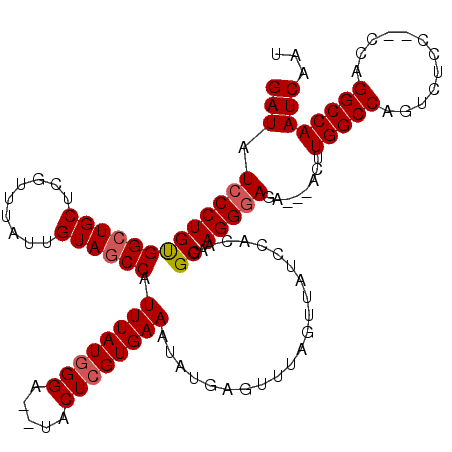
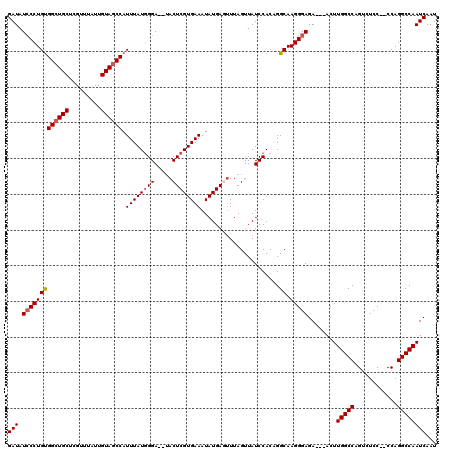
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:28 2006