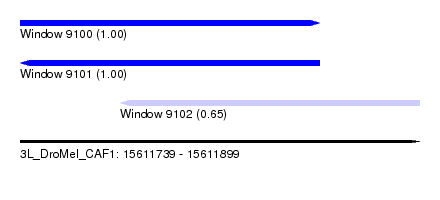
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,611,739 – 15,611,899 |
| Length | 160 |
| Max. P | 0.999770 |

| Location | 15,611,739 – 15,611,859 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -28.70 |
| Energy contribution | -31.10 |
| Covariance contribution | 2.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15611739 120 + 23771897 CCCGACUGGCAUAUUUGUGUGUGAAUGUUUGCGUAGAAGCAUUCAUUUCUGUGAACGUUUCGUUAACUUGGCCAAAAGCCAAUUUUCAGCCAAAACGCAUGCCAAAUUUGCCAAAAACGG .(((..(((((.(((((.(((((....((((.((.(((((.(((((....))))).)))))......(((((.....)))))......))))))...)))))))))).)))))....))) ( -37.00) >DroSec_CAF1 34812 120 + 1 CCCGACUGGCAUAUUUGUGUGUGAAUGUUUGUGUAGAAGCAUUCAUUUCUGUGAACGUUUCGUUAACUUGGCCAAAAGCCAAUUUUCAGCCAAAACGCAUGCCAAAUUUGCCAAAAACGG .(((..(((((.(((((.(((((....((((.((.(((((.(((((....))))).)))))......(((((.....)))))......))))))...)))))))))).)))))....))) ( -37.00) >DroSim_CAF1 34851 120 + 1 CCCGACUGGCAUAUUUGCGUGUGAAUGUUUGUGUAGAAGCAUUCAUUUCUGUGAACGUUUCGUUAACUUGGCCAAAAGCCAAUUUUCAGCCAAAACGCAUGCCAAAUUUGCCAAAAACGG .(((..(((((.(((((((((((....((((.((.(((((.(((((....))))).)))))......(((((.....)))))......)))))).)))))).))))).)))))....))) ( -38.50) >DroEre_CAF1 34906 120 + 1 CCCGUCUGGCAUAUUUGUGUGUGCAUGUUUGUGUAGAAGCAUUCAUUUCUCUGAACGUUUCGUUAACUUGGCCAAAAGCCAAUUUUCAGCCAAAACGCAUGCCAAAUUUGCCAAAAACGG .((((.(((((.(((((.((((((...((((.((.(((((.((((......)))).)))))......(((((.....)))))......))))))..))))))))))).)))))...)))) ( -39.80) >DroYak_CAF1 37268 106 + 1 CCAGUCUGGCAUAUUU--------------GUGUAGAAGCAUUCAUUUCUCUGAACGUUUCGUUAACUUGGCCAAAAGCCAAUUUUCAGCCAAAACGCAUGCCAAAUUUGCCAAAAACGG ......(((((.((((--------------(.((((((((.((((......)))).)))))(((...(((((..((((....))))..))))))))...)))))))).)))))....... ( -27.30) >consensus CCCGACUGGCAUAUUUGUGUGUGAAUGUUUGUGUAGAAGCAUUCAUUUCUGUGAACGUUUCGUUAACUUGGCCAAAAGCCAAUUUUCAGCCAAAACGCAUGCCAAAUUUGCCAAAAACGG .(((..(((((.(((((.(((((....((((.((.(((((.(((((....))))).)))))......(((((.....)))))......))))))...)))))))))).)))))....))) (-28.70 = -31.10 + 2.40)

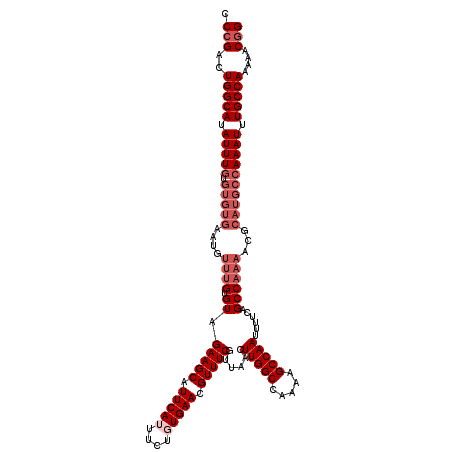
| Location | 15,611,739 – 15,611,859 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -31.96 |
| Energy contribution | -32.36 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.87 |
| SVM decision value | 4.04 |
| SVM RNA-class probability | 0.999770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15611739 120 - 23771897 CCGUUUUUGGCAAAUUUGGCAUGCGUUUUGGCUGAAAAUUGGCUUUUGGCCAAGUUAACGAAACGUUCACAGAAAUGAAUGCUUCUACGCAAACAUUCACACACAAAUAUGCCAGUCGGG (((...((((((.(((((....((((((((..(((...(((((.....))))).))).)))))))).....(((.((..(((......)))..))))).....))))).)))))).))). ( -37.30) >DroSec_CAF1 34812 120 - 1 CCGUUUUUGGCAAAUUUGGCAUGCGUUUUGGCUGAAAAUUGGCUUUUGGCCAAGUUAACGAAACGUUCACAGAAAUGAAUGCUUCUACACAAACAUUCACACACAAAUAUGCCAGUCGGG (((...((((((.(((((....((((((((..(((...(((((.....))))).))).)))))))).........((((((.((......)).))))))....))))).)))))).))). ( -36.70) >DroSim_CAF1 34851 120 - 1 CCGUUUUUGGCAAAUUUGGCAUGCGUUUUGGCUGAAAAUUGGCUUUUGGCCAAGUUAACGAAACGUUCACAGAAAUGAAUGCUUCUACACAAACAUUCACACGCAAAUAUGCCAGUCGGG (((...((((((.(((((.(..((((((((..(((...(((((.....))))).))).)))))))).........((((((.((......)).))))))...)))))).)))))).))). ( -36.50) >DroEre_CAF1 34906 120 - 1 CCGUUUUUGGCAAAUUUGGCAUGCGUUUUGGCUGAAAAUUGGCUUUUGGCCAAGUUAACGAAACGUUCAGAGAAAUGAAUGCUUCUACACAAACAUGCACACACAAAUAUGCCAGACGGG (((((..(((((.((((((..(((((((((((..(((......)))..)))))).....(((.((((((......)))))).))).........)))))..).))))).)))))))))). ( -41.00) >DroYak_CAF1 37268 106 - 1 CCGUUUUUGGCAAAUUUGGCAUGCGUUUUGGCUGAAAAUUGGCUUUUGGCCAAGUUAACGAAACGUUCAGAGAAAUGAAUGCUUCUACAC--------------AAAUAUGCCAGACUGG (((..(((((((.(((((........((((((..(((......)))..)))))).....(((.((((((......)))))).)))....)--------------)))).))))))).))) ( -32.90) >consensus CCGUUUUUGGCAAAUUUGGCAUGCGUUUUGGCUGAAAAUUGGCUUUUGGCCAAGUUAACGAAACGUUCACAGAAAUGAAUGCUUCUACACAAACAUUCACACACAAAUAUGCCAGUCGGG (((...((((((.(((((........((((((..(((......)))..)))))).....(((.((((((......)))))).)))..................))))).)))))).))). (-31.96 = -32.36 + 0.40)

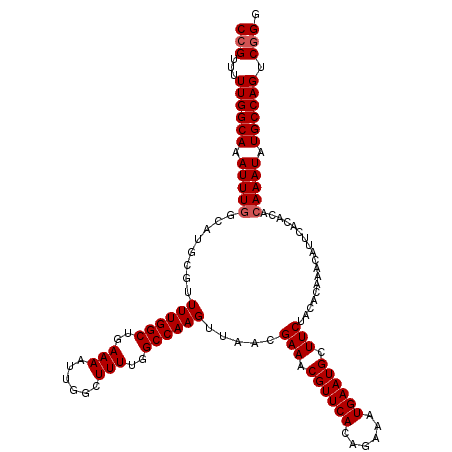

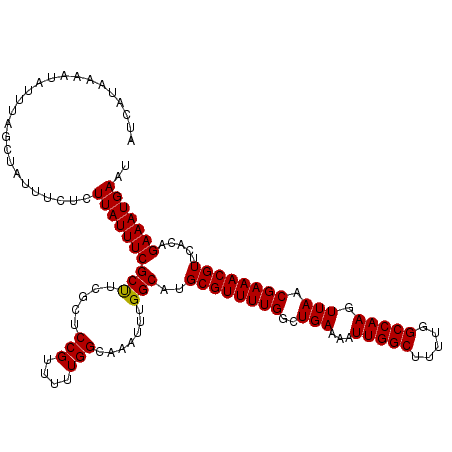
| Location | 15,611,779 – 15,611,899 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -26.00 |
| Energy contribution | -25.84 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15611779 120 - 23771897 AUCAUAAAAUAUUUAGCUAUUUCUCUUAUUUCGCUUCGCUCCGUUUUUGGCAAAUUUGGCAUGCGUUUUGGCUGAAAAUUGGCUUUUGGCCAAGUUAACGAAACGUUCACAGAAAUGAAU .........................((((((((((.....(((....))).......)))..((((((((..(((...(((((.....))))).))).)))))))).....))))))).. ( -25.60) >DroSec_CAF1 34852 120 - 1 AUCAUAAAAUAUUUAGCUAUUUCUCUUAUUUCGCUACGCUCCGUUUUUGGCAAAUUUGGCAUGCGUUUUGGCUGAAAAUUGGCUUUUGGCCAAGUUAACGAAACGUUCACAGAAAUGAAU .........................(((((((((((....(((....)))......))))..((((((((..(((...(((((.....))))).))).)))))))).....))))))).. ( -26.10) >DroSim_CAF1 34891 120 - 1 AUCAUAAAAUAUUUAGCUAUGUCUCUUAUUUCGCUUCGCUCCGUUUUUGGCAAAUUUGGCAUGCGUUUUGGCUGAAAAUUGGCUUUUGGCCAAGUUAACGAAACGUUCACAGAAAUGAAU .........................((((((((((.....(((....))).......)))..((((((((..(((...(((((.....))))).))).)))))))).....))))))).. ( -25.60) >DroEre_CAF1 34946 120 - 1 AUCAUAAAAUAUUUAGCUAUUUCUCUUAUUUCGCCUCGCUCCGUUUUUGGCAAAUUUGGCAUGCGUUUUGGCUGAAAAUUGGCUUUUGGCCAAGUUAACGAAACGUUCAGAGAAAUGAAU .................(((((((((......(((.....(((....))).......)))..((((((((..(((...(((((.....))))).))).))))))))..)))))))))... ( -30.80) >DroYak_CAF1 37294 120 - 1 AUCAUAAAAUAUUUAGCUAUUUCUCUUAUUUCGCUUCGCUCCGUUUUUGGCAAAUUUGGCAUGCGUUUUGGCUGAAAAUUGGCUUUUGGCCAAGUUAACGAAACGUUCAGAGAAAUGAAU .................(((((((((......(((.....(((....))).......)))..((((((((..(((...(((((.....))))).))).))))))))..)))))))))... ( -28.90) >consensus AUCAUAAAAUAUUUAGCUAUUUCUCUUAUUUCGCUUCGCUCCGUUUUUGGCAAAUUUGGCAUGCGUUUUGGCUGAAAAUUGGCUUUUGGCCAAGUUAACGAAACGUUCACAGAAAUGAAU .........................((((((((((.....(((....))).......)))..((((((((..(((...(((((.....))))).))).)))))))).....))))))).. (-26.00 = -25.84 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:25 2006