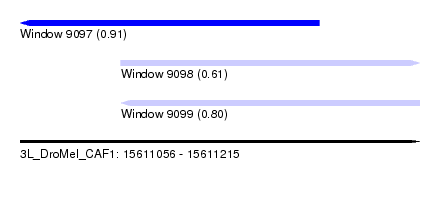
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,611,056 – 15,611,215 |
| Length | 159 |
| Max. P | 0.911249 |

| Location | 15,611,056 – 15,611,175 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.48 |
| Mean single sequence MFE | -31.64 |
| Consensus MFE | -27.28 |
| Energy contribution | -27.76 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
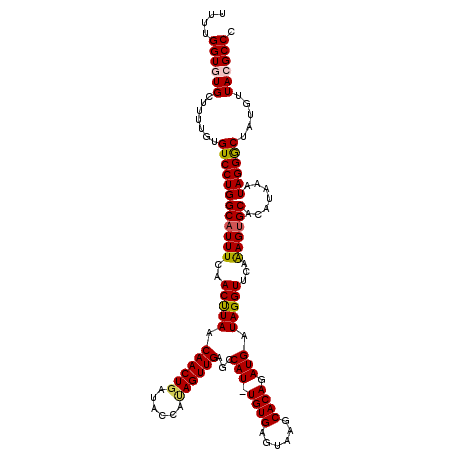
>3L_DroMel_CAF1 15611056 119 - 23771897 CUGAUACCAUAGUUGAGCCAU-UGUGAGUAAGCACAGAUGAUAGGUUCAAAGUGCACAUAAAUAGGGCUAUGUUACGCCCAUUUGAGUGUGCAAAAUUGUGUUUAACUAAAACCGAUGAC .........((((((((((((-((((......))))).(((.....)))..(..(((.(((((.((((........))))))))).)))..).....)).)))))))))........... ( -31.50) >DroSec_CAF1 34157 119 - 1 CUGAUACCAUAGUUGAGCCAU-UGUGAGUAAGCACAGAUGAUAGGUUCAAAGUGCACAUAAAUAGGGCUAUGUUACGCCCAUUUGAGUGUGCAAAAUUGUGUUUAACGAAAAUCGAUGAC ...........(((((....(-(((...(((((((((.(((.....)))..(..(((.(((((.((((........))))))))).)))..)....)))))))))))))...)))))... ( -31.60) >DroSim_CAF1 34169 119 - 1 CUGAUACCAUAGUUGAGCCAU-UGUGAGUAAGCACAGAUGAUAGGUUCAAAGUGCACAUAAAUAGGGCUAUGUUACGCCCAUUUGAGUGUGCAAAAUUGUGUUUAACGAAAACCGAUGAC ............(((((((.(-((((......)))))......))))))).(..(((.(((((.((((........))))))))).)))..)...((((.((((.....))))))))... ( -30.50) >DroEre_CAF1 34222 119 - 1 CUGAUACCAAAGUUGAGCCAUUUGUGAGUAAGCACAGAUGAUAGGUUCAAAGUGCACAUAAAUAGGGCUGUGUUACGCCCAUUUGGCUGGGCAAAAUUGUGUUUAACUAAG-GCGAAGAC ......((..((((((((((((((((......))))))))...........((.((..(((((.((((........)))))))))..)).))........))))))))..)-)....... ( -31.10) >DroYak_CAF1 36485 119 - 1 CUGAUACCACAGUUGAGCCAUUUGUGAGUAAGCACAGAUGAUAGGUUCGGAGUGCACAUAAAUAGGACUCUGUUACGCCCAUUUGGGUGUGCAAAAUUGUGUUUAACUAAA-GCGAUGAA .......(((((((..((((((((((......)))))))).......((((((.(.........).))))))..((((((....))))))))..)))))))..........-........ ( -33.50) >consensus CUGAUACCAUAGUUGAGCCAU_UGUGAGUAAGCACAGAUGAUAGGUUCAAAGUGCACAUAAAUAGGGCUAUGUUACGCCCAUUUGAGUGUGCAAAAUUGUGUUUAACUAAAACCGAUGAC .......((((((((((((((.((((......))))....)).)))))...(..(((.(((((.((((........))))))))).)))..)..)))))))................... (-27.28 = -27.76 + 0.48)



| Location | 15,611,096 – 15,611,215 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.49 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -25.80 |
| Energy contribution | -25.68 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15611096 119 + 23771897 GGGCGUAACAUAGCCCUAUUUAUGUGCACUUUGAACCUAUCAUCUGUGCUUACUCACA-AUGGCUCAACUAUGGUAUCAGUUGUUAAGUUGAAAUGCCAGGACACAAAAGCACACCAAAA ((((........))))......(((((..((((..(((......((((......))))-..(((((((((.(..((.....))..)))))))...))))))...)))).)))))...... ( -28.90) >DroSec_CAF1 34197 119 + 1 GGGCGUAACAUAGCCCUAUUUAUGUGCACUUUGAACCUAUCAUCUGUGCUUACUCACA-AUGGCUCAACUAUGGUAUCAGUUGUUAGGUUGAAAUGCCAGGACACAAAAGCAUACCAAAA ((((........))))....((((((((.(((.((((((.((.(((....((((....-((((.....)))))))).))).)).)))))).))))))..(....)....)))))...... ( -27.60) >DroSim_CAF1 34209 119 + 1 GGGCGUAACAUAGCCCUAUUUAUGUGCACUUUGAACCUAUCAUCUGUGCUUACUCACA-AUGGCUCAACUAUGGUAUCAGUUGUUAAGUUGAAAUGCCAGGACACAAAAGCAUACCAAAA ((((........))))....(((((((((..(((.....)))...))))....((...-.((((((((((.(..((.....))..)))))))...)))).)).......)))))...... ( -27.70) >DroEre_CAF1 34261 120 + 1 GGGCGUAACACAGCCCUAUUUAUGUGCACUUUGAACCUAUCAUCUGUGCUUACUCACAAAUGGCUCAACUUUGGUAUCAGUUGUUAAGUGGAAAGGCCAGGACACAAAAGCACACCAAAA ((((........))))......(((((....(((.....)))((((.((((..((((.((..(((..((....))...)))..))..))))..))))))))........)))))...... ( -29.30) >DroYak_CAF1 36524 120 + 1 GGGCGUAACAGAGUCCUAUUUAUGUGCACUCCGAACCUAUCAUCUGUGCUUACUCACAAAUGGCUCAACUGUGGUAUCAGUUGUUAAGUUGAAAUGCCAGGACACAAGAGCACACCAAAA ((((........))))......(((((.((.....(((......((((......))))...(((.((((((......))))))............)))))).....)).)))))...... ( -26.42) >consensus GGGCGUAACAUAGCCCUAUUUAUGUGCACUUUGAACCUAUCAUCUGUGCUUACUCACA_AUGGCUCAACUAUGGUAUCAGUUGUUAAGUUGAAAUGCCAGGACACAAAAGCACACCAAAA ((((........))))......(((((..((((..(((......((((......))))...(((((((((.(..((.....))..)))))))...))))))...)))).)))))...... (-25.80 = -25.68 + -0.12)



| Location | 15,611,096 – 15,611,215 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.49 |
| Mean single sequence MFE | -33.26 |
| Consensus MFE | -27.93 |
| Energy contribution | -28.13 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15611096 119 - 23771897 UUUUGGUGUGCUUUUGUGUCCUGGCAUUUCAACUUAACAACUGAUACCAUAGUUGAGCCAU-UGUGAGUAAGCACAGAUGAUAGGUUCAAAGUGCACAUAAAUAGGGCUAUGUUACGCCC .....(((((((((((...((((((...((((((................))))))))).(-((((......))))).....)))..))))).)))))).....((((........)))) ( -32.59) >DroSec_CAF1 34197 119 - 1 UUUUGGUAUGCUUUUGUGUCCUGGCAUUUCAACCUAACAACUGAUACCAUAGUUGAGCCAU-UGUGAGUAAGCACAGAUGAUAGGUUCAAAGUGCACAUAAAUAGGGCUAUGUUACGCCC ....((.((((....))))))((((((((.((((((.((.(((.(((((((((......))-)))).)))....))).)).))))))..)))))).))......((((........)))) ( -29.70) >DroSim_CAF1 34209 119 - 1 UUUUGGUAUGCUUUUGUGUCCUGGCAUUUCAACUUAACAACUGAUACCAUAGUUGAGCCAU-UGUGAGUAAGCACAGAUGAUAGGUUCAAAGUGCACAUAAAUAGGGCUAUGUUACGCCC ....((.((((....))))))((((((((.((((((.((.(((.(((((((((......))-)))).)))....))).)).))))))..)))))).))......((((........)))) ( -27.00) >DroEre_CAF1 34261 120 - 1 UUUUGGUGUGCUUUUGUGUCCUGGCCUUUCCACUUAACAACUGAUACCAAAGUUGAGCCAUUUGUGAGUAAGCACAGAUGAUAGGUUCAAAGUGCACAUAAAUAGGGCUGUGUUACGCCC .....(((((((((((...((((..............(((((........)))))...((((((((......)))))))).))))..))))).)))))).....((((........)))) ( -35.60) >DroYak_CAF1 36524 120 - 1 UUUUGGUGUGCUCUUGUGUCCUGGCAUUUCAACUUAACAACUGAUACCACAGUUGAGCCAUUUGUGAGUAAGCACAGAUGAUAGGUUCGGAGUGCACAUAAAUAGGACUCUGUUACGCCC ....(((((((....(.(((((((((((((((((((.((((((......))))))...((((((((......)))))))).)))))).))))))).......)))))).).).)))))). ( -41.41) >consensus UUUUGGUGUGCUUUUGUGUCCUGGCAUUUCAACUUAACAACUGAUACCAUAGUUGAGCCAU_UGUGAGUAAGCACAGAUGAUAGGUUCAAAGUGCACAUAAAUAGGGCUAUGUUACGCCC ....((((((.......((((((((((((..(((((.((((((......))))))...(((.((((......)))).))).)))))...)))))).......)))))).....)))))). (-27.93 = -28.13 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:22 2006