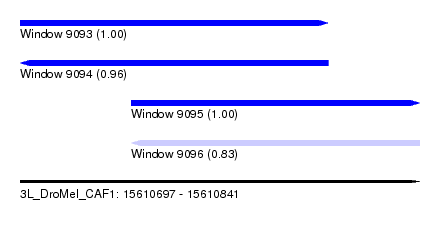
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,610,697 – 15,610,841 |
| Length | 144 |
| Max. P | 0.999883 |

| Location | 15,610,697 – 15,610,808 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.19 |
| Mean single sequence MFE | -33.67 |
| Consensus MFE | -31.33 |
| Energy contribution | -31.53 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.37 |
| SVM RNA-class probability | 0.999883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15610697 111 + 23771897 GGCCACAGUUCGAAACGAAAGUAGAAGAAUUUAUUGCACUGACAUACUAAGAUGCU---------UCCUAUUUCGUUAUCGAAAUGGGCCAAAUGCGCUCGAGCGUAUAAAUUGAUUGUG ((((.((.((((((((((((...((((.((((.................)))).))---------))...))))))).))))).))))))..((((((....))))))............ ( -32.63) >DroSec_CAF1 33797 111 + 1 GGCCACAGUUCGAAACGAAAGUGGAGGAAUUUAUUGCACUGACAUACUAAGAUGCU---------UCCUAUUUCGUUAUCGAAAUGGGCCAAAUGCGCUCGAGCGUAUAAAUUGAUUGGG ((((.((.((((((((((((....(((((......(((((.........)).))))---------)))).))))))).))))).))))))..((((((....))))))............ ( -35.80) >DroSim_CAF1 33813 111 + 1 GGCCACAGUUCGAAACGAAAGUGGAGGAAUUUAUUGCACUGACAUACUAAGAUGCU---------UCCUAUUUCGUUAUCGAAAUGGGCCAAAUGCGCUCGAGCGUAUAAAUUGAUUGUG ((((.((.((((((((((((....(((((......(((((.........)).))))---------)))).))))))).))))).))))))..((((((....))))))............ ( -35.80) >DroEre_CAF1 33882 109 + 1 GGCC--AGUUCGAAACGAAAGUGGAAGAAUUUAUUGCACUGACAUACUAAGAUGCU---------UCCUAUUUUGUUAUCGAAAUGGGGCAAGUGCGCUCGAGCGUAUAAAUUGAAUGUG .(((--..((((((((((((..(((((.((((.................)))).))---------)))..))))))).)))))....)))..((((((....))))))............ ( -29.23) >DroYak_CAF1 36096 120 + 1 GGCCACAGUUCGAAACGAAAGUGGAAGAAUUUAUUGCACUGACAUACUAAGAUGCUAUUCUGCUAUCCUAUUUUGUUAUCGAAAUGGGCCAAAUGCGCUCGAGCGUAUAAAUUGAAUGUG ((((.((.((((((((((((..((((((((.....(((((.........)).))).)))))....)))..))))))).))))).))))))..((((((....))))))............ ( -34.90) >consensus GGCCACAGUUCGAAACGAAAGUGGAAGAAUUUAUUGCACUGACAUACUAAGAUGCU_________UCCUAUUUCGUUAUCGAAAUGGGCCAAAUGCGCUCGAGCGUAUAAAUUGAUUGUG ((((.((.((((((((((((..(((..........(((((.........)).)))..........)))..))))))).))))).))))))..((((((....))))))............ (-31.33 = -31.53 + 0.20)


| Location | 15,610,697 – 15,610,808 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.19 |
| Mean single sequence MFE | -26.14 |
| Consensus MFE | -22.77 |
| Energy contribution | -23.41 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15610697 111 - 23771897 CACAAUCAAUUUAUACGCUCGAGCGCAUUUGGCCCAUUUCGAUAACGAAAUAGGA---------AGCAUCUUAGUAUGUCAGUGCAAUAAAUUCUUCUACUUUCGUUUCGAACUGUGGCC ...............(((....))).....((((((.(((((.(((((((..(((---------((.......((((....))))........)))))..)))))))))))).)).)))) ( -28.66) >DroSec_CAF1 33797 111 - 1 CCCAAUCAAUUUAUACGCUCGAGCGCAUUUGGCCCAUUUCGAUAACGAAAUAGGA---------AGCAUCUUAGUAUGUCAGUGCAAUAAAUUCCUCCACUUUCGUUUCGAACUGUGGCC ...............(((....))).....((((((.(((((.(((((((.((((---------(........((((....))))......)))))....)))))))))))).)).)))) ( -28.24) >DroSim_CAF1 33813 111 - 1 CACAAUCAAUUUAUACGCUCGAGCGCAUUUGGCCCAUUUCGAUAACGAAAUAGGA---------AGCAUCUUAGUAUGUCAGUGCAAUAAAUUCCUCCACUUUCGUUUCGAACUGUGGCC ...............(((....))).....((((((.(((((.(((((((.((((---------(........((((....))))......)))))....)))))))))))).)).)))) ( -28.24) >DroEre_CAF1 33882 109 - 1 CACAUUCAAUUUAUACGCUCGAGCGCACUUGCCCCAUUUCGAUAACAAAAUAGGA---------AGCAUCUUAGUAUGUCAGUGCAAUAAAUUCUUCCACUUUCGUUUCGAACU--GGCC ...............(((....)))........(((.(((((.(((.(((..(((---------((.......((((....))))........)))))..))).)))))))).)--)).. ( -19.26) >DroYak_CAF1 36096 120 - 1 CACAUUCAAUUUAUACGCUCGAGCGCAUUUGGCCCAUUUCGAUAACAAAAUAGGAUAGCAGAAUAGCAUCUUAGUAUGUCAGUGCAAUAAAUUCUUCCACUUUCGUUUCGAACUGUGGCC ...............(((....))).....((((((.(((((.(((....((((((.((......))))))))((((....))))...................)))))))).)).)))) ( -26.30) >consensus CACAAUCAAUUUAUACGCUCGAGCGCAUUUGGCCCAUUUCGAUAACGAAAUAGGA_________AGCAUCUUAGUAUGUCAGUGCAAUAAAUUCUUCCACUUUCGUUUCGAACUGUGGCC ...............(((....))).....((((((.(((((.(((((((..(((..........((((............))))..........)))..)))))))))))).)).)))) (-22.77 = -23.41 + 0.64)

| Location | 15,610,737 – 15,610,841 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.58 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -28.78 |
| Energy contribution | -28.46 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.49 |
| SVM RNA-class probability | 0.999298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15610737 104 + 23771897 GACAUACUAAGAUGCU---------UCCUAUUUCGUUAUCGAAAUGGGCCAAAUGCGCUCGAGCGUAUAAAUUGAUUGUGCCCCACCA-------UGAGGUAGGUACAGAGAAAUAUGUU ((((((..........---------.(((((((((....)))))))))....((((((....)))))).......(((((((..(((.-------...))).))))))).....)))))) ( -31.50) >DroSec_CAF1 33837 104 + 1 GACAUACUAAGAUGCU---------UCCUAUUUCGUUAUCGAAAUGGGCCAAAUGCGCUCGAGCGUAUAAAUUGAUUGGGCCCCACCA-------UGAGGUAGGUACAAAGAAAUAUGUU ((((((..........---------.(((((((((....)))))))))....((((((....)))))).......(((..((..(((.-------...))).))..))).....)))))) ( -28.00) >DroSim_CAF1 33853 104 + 1 GACAUACUAAGAUGCU---------UCCUAUUUCGUUAUCGAAAUGGGCCAAAUGCGCUCGAGCGUAUAAAUUGAUUGUGCCCCACCA-------UGAGGUAGGUACAAAGAAAUAUGUU ((((((..........---------.(((((((((....)))))))))....((((((....)))))).......(((((((..(((.-------...))).))))))).....)))))) ( -31.80) >DroEre_CAF1 33920 104 + 1 GACAUACUAAGAUGCU---------UCCUAUUUUGUUAUCGAAAUGGGGCAAGUGCGCUCGAGCGUAUAAAUUGAAUGUGCCCCACCA-------AGAGGUAGGUACAAAGAAAUAUGUU ((((((......(((.---------.(((((((((....)))))))))))).((((((....))))))........((((((..(((.-------...))).))))))......)))))) ( -31.90) >DroYak_CAF1 36136 120 + 1 GACAUACUAAGAUGCUAUUCUGCUAUCCUAUUUUGUUAUCGAAAUGGGCCAAAUGCGCUCGAGCGUAUAAAUUGAAUGUGCCCCACCAUGCGCCAUGUGGUAGGUGCAAAGAAAUAUGUU ((((((.........(((((......(((((((((....)))))))))....((((((....)))))).....)))))(((((.(((((.......))))).)).)))......)))))) ( -32.30) >consensus GACAUACUAAGAUGCU_________UCCUAUUUCGUUAUCGAAAUGGGCCAAAUGCGCUCGAGCGUAUAAAUUGAUUGUGCCCCACCA_______UGAGGUAGGUACAAAGAAAUAUGUU ((((((....................(((((((((....)))))))))....((((((....))))))........((((((..(((...........))).))))))......)))))) (-28.78 = -28.46 + -0.32)



| Location | 15,610,737 – 15,610,841 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.58 |
| Mean single sequence MFE | -25.55 |
| Consensus MFE | -23.14 |
| Energy contribution | -23.38 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.825042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15610737 104 - 23771897 AACAUAUUUCUCUGUACCUACCUCA-------UGGUGGGGCACAAUCAAUUUAUACGCUCGAGCGCAUUUGGCCCAUUUCGAUAACGAAAUAGGA---------AGCAUCUUAGUAUGUC .(((((((........((((((...-------.))))))((..............(((....))).......((.((((((....)))))).)).---------.)).....))))))). ( -25.80) >DroSec_CAF1 33837 104 - 1 AACAUAUUUCUUUGUACCUACCUCA-------UGGUGGGGCCCAAUCAAUUUAUACGCUCGAGCGCAUUUGGCCCAUUUCGAUAACGAAAUAGGA---------AGCAUCUUAGUAUGUC .(((((((....(((.((((((...-------.))))))(((.(((.........(((....))).))).)))((((((((....)))))).)).---------.)))....))))))). ( -26.80) >DroSim_CAF1 33853 104 - 1 AACAUAUUUCUUUGUACCUACCUCA-------UGGUGGGGCACAAUCAAUUUAUACGCUCGAGCGCAUUUGGCCCAUUUCGAUAACGAAAUAGGA---------AGCAUCUUAGUAUGUC .(((((((....(((.((((((...-------.))))))................(((....))).......((.((((((....)))))).)).---------.)))....))))))). ( -26.00) >DroEre_CAF1 33920 104 - 1 AACAUAUUUCUUUGUACCUACCUCU-------UGGUGGGGCACAUUCAAUUUAUACGCUCGAGCGCACUUGCCCCAUUUCGAUAACAAAAUAGGA---------AGCAUCUUAGUAUGUC .(((((((....(((.((((((...-------.))(((((((.............(((....)))....)))))))..............)))).---------.)))....))))))). ( -23.83) >DroYak_CAF1 36136 120 - 1 AACAUAUUUCUUUGCACCUACCACAUGGCGCAUGGUGGGGCACAUUCAAUUUAUACGCUCGAGCGCAUUUGGCCCAUUUCGAUAACAAAAUAGGAUAGCAGAAUAGCAUCUUAGUAUGUC .((((((..((.(((.((........)).))).))..((((.((........((.(((....))).)).))))))...............((((((.((......)))))))))))))). ( -25.30) >consensus AACAUAUUUCUUUGUACCUACCUCA_______UGGUGGGGCACAAUCAAUUUAUACGCUCGAGCGCAUUUGGCCCAUUUCGAUAACGAAAUAGGA_________AGCAUCUUAGUAUGUC .(((((((....(((.((((((...........))))))................(((....))).......((.((((((....)))))).))...........)))....))))))). (-23.14 = -23.38 + 0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:19 2006