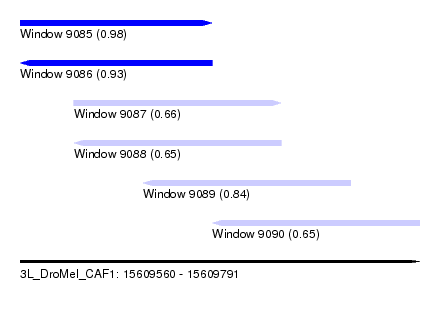
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,609,560 – 15,609,791 |
| Length | 231 |
| Max. P | 0.977576 |

| Location | 15,609,560 – 15,609,671 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.31 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -21.44 |
| Energy contribution | -21.28 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15609560 111 + 23771897 GAGCCAC---------CACGCACACGCACAAUGAAUAAUUUAAGGACCCCGAAGACAGACGGGUCCUGGCAUCAUCAAAUAAAUGCCAAAUGAGCAAUCAAAGUGCCAUGAAAGACUAAC ..((...---------...))....((((.............(((((((...........)))))))(((((..........)))))...(((....)))..)))).............. ( -25.30) >DroSec_CAF1 32643 111 + 1 GAGCCAC---------CACGCACACGCACAAUGAAUAAUUUAAGGACCCCGAAGACAGACGGGUCCUGGCAUCAUCAAAUAAAUGCCAAAUGAGCAAUCAAAGUGCCAUGAAAGACUAAC ..((...---------...))....((((.............(((((((...........)))))))(((((..........)))))...(((....)))..)))).............. ( -25.30) >DroSim_CAF1 32671 111 + 1 GAGCCAC---------CACGCACACGCACAAUGAAUAAUUUAAGGACCCCGAAGACAGACGGGUCCUGGCAUCAUCAAAUAAAUGCCAAAUGAGCAAUCAAAGUGCCAUGAAAGACUAAC ..((...---------...))....((((.............(((((((...........)))))))(((((..........)))))...(((....)))..)))).............. ( -25.30) >DroEre_CAF1 32772 101 + 1 -------------------GCACACGCACAGUGAAUAAUUUAAGGACUCCGUAGACAGACGGAUCCUGGCAUCAUCAAAUAAAUGCCAAAUGAGCAAUCAAAGUGCCAUGAAAGACUAAC -------------------......((((..(((........((((.(((((......)))))))))(((((..........)))))..........)))..)))).............. ( -24.90) >DroYak_CAF1 34927 120 + 1 GAGCCAAGCACACGCACACGCACACGCACAAUGAAUAAUUUAAGGACUCCGUAGACAGAGGGGCCUUGGCAUCAUCAAAUAAAUGCCAAAUGAGCAAUCAAAGUGCCAUGAAAGACUAAC .......((((..((....))....((.((...........((((.((((.........))))))))(((((..........)))))...)).)).......)))).............. ( -24.70) >consensus GAGCCAC_________CACGCACACGCACAAUGAAUAAUUUAAGGACCCCGAAGACAGACGGGUCCUGGCAUCAUCAAAUAAAUGCCAAAUGAGCAAUCAAAGUGCCAUGAAAGACUAAC .........................((((..(((........((((.((((........))))))))(((((..........)))))..........)))..)))).............. (-21.44 = -21.28 + -0.16)



| Location | 15,609,560 – 15,609,671 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.31 |
| Mean single sequence MFE | -32.84 |
| Consensus MFE | -26.58 |
| Energy contribution | -26.66 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15609560 111 - 23771897 GUUAGUCUUUCAUGGCACUUUGAUUGCUCAUUUGGCAUUUAUUUGAUGAUGCCAGGACCCGUCUGUCUUCGGGGUCCUUAAAUUAUUCAUUGUGCGUGUGCGUG---------GUGGCUC ...((((..(((((((((..(((.........(((((((........)))))))((((((...........)))))).........)))..)))).....))))---------).)))). ( -34.00) >DroSec_CAF1 32643 111 - 1 GUUAGUCUUUCAUGGCACUUUGAUUGCUCAUUUGGCAUUUAUUUGAUGAUGCCAGGACCCGUCUGUCUUCGGGGUCCUUAAAUUAUUCAUUGUGCGUGUGCGUG---------GUGGCUC ...((((..(((((((((..(((.........(((((((........)))))))((((((...........)))))).........)))..)))).....))))---------).)))). ( -34.00) >DroSim_CAF1 32671 111 - 1 GUUAGUCUUUCAUGGCACUUUGAUUGCUCAUUUGGCAUUUAUUUGAUGAUGCCAGGACCCGUCUGUCUUCGGGGUCCUUAAAUUAUUCAUUGUGCGUGUGCGUG---------GUGGCUC ...((((..(((((((((..(((.........(((((((........)))))))((((((...........)))))).........)))..)))).....))))---------).)))). ( -34.00) >DroEre_CAF1 32772 101 - 1 GUUAGUCUUUCAUGGCACUUUGAUUGCUCAUUUGGCAUUUAUUUGAUGAUGCCAGGAUCCGUCUGUCUACGGAGUCCUUAAAUUAUUCACUGUGCGUGUGC------------------- ..........((((((((..(((.........(((((((........)))))))((((((((......))))).))).........)))..)))).)))).------------------- ( -27.30) >DroYak_CAF1 34927 120 - 1 GUUAGUCUUUCAUGGCACUUUGAUUGCUCAUUUGGCAUUUAUUUGAUGAUGCCAAGGCCCCUCUGUCUACGGAGUCCUUAAAUUAUUCAUUGUGCGUGUGCGUGUGCGUGUGCUUGGCUC ...((((.......((((..(((..(((...((((((((........)))))))))))..(((((....)))))............)))..))))(((..((....))..)))..)))). ( -34.90) >consensus GUUAGUCUUUCAUGGCACUUUGAUUGCUCAUUUGGCAUUUAUUUGAUGAUGCCAGGACCCGUCUGUCUUCGGGGUCCUUAAAUUAUUCAUUGUGCGUGUGCGUG_________GUGGCUC ..........((((((((..(((.........(((((((........)))))))(((((((........))).)))).........)))..)))).)))).................... (-26.58 = -26.66 + 0.08)



| Location | 15,609,591 – 15,609,711 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -23.44 |
| Energy contribution | -23.48 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15609591 120 + 23771897 UAAGGACCCCGAAGACAGACGGGUCCUGGCAUCAUCAAAUAAAUGCCAAAUGAGCAAUCAAAGUGCCAUGAAAGACUAACUAAGCAGAGCCACCAACUGAUGAGGGGCUGUAAAAAAUGA ..(((((((...........)))))))(((((..........)))))..(((.(((.......))))))................(.((((.((.........)))))).)......... ( -27.40) >DroSec_CAF1 32674 120 + 1 UAAGGACCCCGAAGACAGACGGGUCCUGGCAUCAUCAAAUAAAUGCCAAAUGAGCAAUCAAAGUGCCAUGAAAGACUAACUAAGCAGAGCCACCAACUGAUGAGGGGCUGUAAAAAAUGA ..(((((((...........)))))))(((((..........)))))..(((.(((.......))))))................(.((((.((.........)))))).)......... ( -27.40) >DroSim_CAF1 32702 120 + 1 UAAGGACCCCGAAGACAGACGGGUCCUGGCAUCAUCAAAUAAAUGCCAAAUGAGCAAUCAAAGUGCCAUGAAAGACUAACUAAGCAGCGCCACCAACUGAUGAGGGGCUGUAAAAAAUGA ..(((((((...........)))))))(((((..........)))))..(((.(((.......))))))..............(((((.((..((.....))..)))))))......... ( -31.00) >DroEre_CAF1 32793 119 + 1 UAAGGACUCCGUAGACAGACGGAUCCUGGCAUCAUCAAAUAAAUGCCAAAUGAGCAAUCAAAGUGCCAUGAAAGACUAACUAAGCAGAGCCACCAUCUGAUG-GGGGCUGUAAAAAAUGA ..((((.(((((......)))))))))(((((..........)))))..(((.(((.......))))))................(.((((.((((...)))-).)))).)......... ( -29.10) >DroYak_CAF1 34967 120 + 1 UAAGGACUCCGUAGACAGAGGGGCCUUGGCAUCAUCAAAUAAAUGCCAAAUGAGCAAUCAAAGUGCCAUGAAAGACUAACUAAGCAGAGCCACCAUCUGAUGAGGGGCUGUAAAAAAUGA ...((.((((.....(((((((((.(((((((..........)))))))(((.(((.......))))))...................))).)).))))....))))))........... ( -28.20) >consensus UAAGGACCCCGAAGACAGACGGGUCCUGGCAUCAUCAAAUAAAUGCCAAAUGAGCAAUCAAAGUGCCAUGAAAGACUAACUAAGCAGAGCCACCAACUGAUGAGGGGCUGUAAAAAAUGA ..((((.((((........))))))))(((((..........)))))..(((.(((.......))))))................(.((((.((.........)))))).)......... (-23.44 = -23.48 + 0.04)

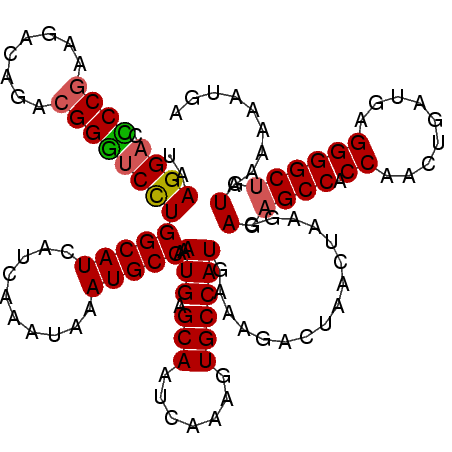

| Location | 15,609,591 – 15,609,711 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -26.74 |
| Energy contribution | -26.66 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.646910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15609591 120 - 23771897 UCAUUUUUUACAGCCCCUCAUCAGUUGGUGGCUCUGCUUAGUUAGUCUUUCAUGGCACUUUGAUUGCUCAUUUGGCAUUUAUUUGAUGAUGCCAGGACCCGUCUGUCUUCGGGGUCCUUA ............(((((....(((.(((.(((....(..(((..(((......))))))..)...)))..(((((((((........)))))))))..))).))).....)))))..... ( -30.10) >DroSec_CAF1 32674 120 - 1 UCAUUUUUUACAGCCCCUCAUCAGUUGGUGGCUCUGCUUAGUUAGUCUUUCAUGGCACUUUGAUUGCUCAUUUGGCAUUUAUUUGAUGAUGCCAGGACCCGUCUGUCUUCGGGGUCCUUA ............(((((....(((.(((.(((....(..(((..(((......))))))..)...)))..(((((((((........)))))))))..))).))).....)))))..... ( -30.10) >DroSim_CAF1 32702 120 - 1 UCAUUUUUUACAGCCCCUCAUCAGUUGGUGGCGCUGCUUAGUUAGUCUUUCAUGGCACUUUGAUUGCUCAUUUGGCAUUUAUUUGAUGAUGCCAGGACCCGUCUGUCUUCGGGGUCCUUA ............(((((.........)).)))((..(..(((..(((......))))))..)...)).....(((((((........)))))))((((((...........))))))... ( -32.60) >DroEre_CAF1 32793 119 - 1 UCAUUUUUUACAGCCCC-CAUCAGAUGGUGGCUCUGCUUAGUUAGUCUUUCAUGGCACUUUGAUUGCUCAUUUGGCAUUUAUUUGAUGAUGCCAGGAUCCGUCUGUCUACGGAGUCCUUA (((........((((.(-(((...)))).))))...........(((......)))....))).........(((((((........)))))))((((((((......))))).)))... ( -29.60) >DroYak_CAF1 34967 120 - 1 UCAUUUUUUACAGCCCCUCAUCAGAUGGUGGCUCUGCUUAGUUAGUCUUUCAUGGCACUUUGAUUGCUCAUUUGGCAUUUAUUUGAUGAUGCCAAGGCCCCUCUGUCUACGGAGUCCUUA ...........((((((((....)).)).))))....................(((....(((....)))(((((((((........)))))))))))).(((((....)))))...... ( -28.00) >consensus UCAUUUUUUACAGCCCCUCAUCAGUUGGUGGCUCUGCUUAGUUAGUCUUUCAUGGCACUUUGAUUGCUCAUUUGGCAUUUAUUUGAUGAUGCCAGGACCCGUCUGUCUUCGGGGUCCUUA ............(((((....(((..((((((....(..(((..(((......))))))..)...)))..(((((((((........))))))))))))...))).....)))))..... (-26.74 = -26.66 + -0.08)



| Location | 15,609,631 – 15,609,751 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -22.06 |
| Energy contribution | -23.06 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15609631 120 - 23771897 ACCUCCCAAAGCCACCCACUGUCCCGAUAAUUCGUGUACAUCAUUUUUUACAGCCCCUCAUCAGUUGGUGGCUCUGCUUAGUUAGUCUUUCAUGGCACUUUGAUUGCUCAUUUGGCAUUU .........(((((((.((((...(((....)))((((.(......).)))).........)))).))))))).((((.(((.(((...(((........)))..))).))).))))... ( -27.00) >DroSec_CAF1 32714 120 - 1 ACCUCCCACAGCCACCCACUGUCCCGAUAAUUCGUGUACAUCAUUUUUUACAGCCCCUCAUCAGUUGGUGGCUCUGCUUAGUUAGUCUUUCAUGGCACUUUGAUUGCUCAUUUGGCAUUU .........(((((((.((((...(((....)))((((.(......).)))).........)))).))))))).((((.(((.(((...(((........)))..))).))).))))... ( -27.00) >DroSim_CAF1 32742 120 - 1 GCCUCCCACAGCCACCCACUGUCCCGAUAAUUCGUGUACAUCAUUUUUUACAGCCCCUCAUCAGUUGGUGGCGCUGCUUAGUUAGUCUUUCAUGGCACUUUGAUUGCUCAUUUGGCAUUU (((.......((((((.((((...(((....)))((((.(......).)))).........)))).))))))((..(..(((..(((......))))))..)...))......))).... ( -29.60) >DroEre_CAF1 32833 117 - 1 --CUCCCAUAGCCACCCACUGUCCCGAUAAUUCGUGUACAUCAUUUUUUACAGCCCC-CAUCAGAUGGUGGCUCUGCUUAGUUAGUCUUUCAUGGCACUUUGAUUGCUCAUUUGGCAUUU --.......(((((((..(((...(((....)))((((.(......).)))).....-...)))..))))))).((((.(((.(((...(((........)))..))).))).))))... ( -23.80) >DroYak_CAF1 35007 120 - 1 ACCUCCCACAAACACCCACCGUCCCGAUAAUUCGUGUACAUCAUUUUUUACAGCCCCUCAUCAGAUGGUGGCUCUGCUUAGUUAGUCUUUCAUGGCACUUUGAUUGCUCAUUUGGCAUUU ...............((((((((.(((....)))((((.(......).))))...........))))))))...((((.(((.(((...(((........)))..))).))).))))... ( -22.50) >consensus ACCUCCCACAGCCACCCACUGUCCCGAUAAUUCGUGUACAUCAUUUUUUACAGCCCCUCAUCAGUUGGUGGCUCUGCUUAGUUAGUCUUUCAUGGCACUUUGAUUGCUCAUUUGGCAUUU ..........((((((.((((...(((....)))((((.(......).)))).........)))).))))))..((((.(((.(((...(((........)))..))).))).))))... (-22.06 = -23.06 + 1.00)

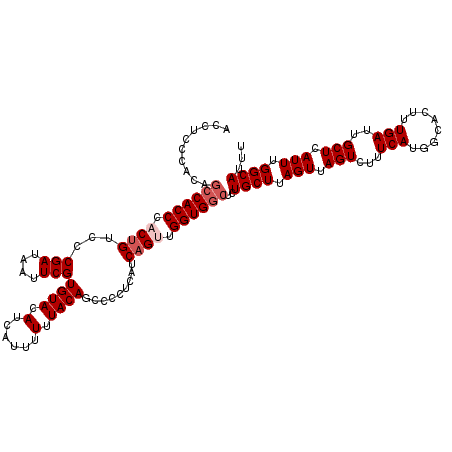
| Location | 15,609,671 – 15,609,791 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.31 |
| Mean single sequence MFE | -20.74 |
| Consensus MFE | -15.10 |
| Energy contribution | -16.30 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15609671 120 - 23771897 GUGAACCGAAACACCGCCCACUCAGCUUGAAACCACCCACACCUCCCAAAGCCACCCACUGUCCCGAUAAUUCGUGUACAUCAUUUUUUACAGCCCCUCAUCAGUUGGUGGCUCUGCUUA (((...((......))..))).........................((.(((((((.((((...(((....)))((((.(......).)))).........)))).))))))).)).... ( -21.90) >DroSec_CAF1 32754 120 - 1 GGGAACCAAAACACCGCCCACUCAGCUCGAAACCACCCACACCUCCCACAGCCACCCACUGUCCCGAUAAUUCGUGUACAUCAUUUUUUACAGCCCCUCAUCAGUUGGUGGCUCUGCUUA ((((...........((.......))..(....).........))))..(((((((.((((...(((....)))((((.(......).)))).........)))).)))))))....... ( -23.40) >DroSim_CAF1 32782 101 - 1 GGGAACCAAAACACC-------------------AAACACGCCUCCCACAGCCACCCACUGUCCCGAUAAUUCGUGUACAUCAUUUUUUACAGCCCCUCAUCAGUUGGUGGCGCUGCUUA ((((...........-------------------.........))))...((((((.((((...(((....)))((((.(......).)))).........)))).))))))........ ( -22.85) >DroEre_CAF1 32873 90 - 1 G-GAACCAGAACAC----------------------------CUCCCAUAGCCACCCACUGUCCCGAUAAUUCGUGUACAUCAUUUUUUACAGCCCC-CAUCAGAUGGUGGCUCUGCUUA (-((..........----------------------------.)))((.(((((((..(((...(((....)))((((.(......).)))).....-...)))..))))))).)).... ( -17.50) >DroYak_CAF1 35047 110 - 1 GGGAAUCAAAACACCGCCCAC----------AACACCCAUACCUCCCACAAACACCCACCGUCCCGAUAAUUCGUGUACAUCAUUUUUUACAGCCCCUCAUCAGAUGGUGGCUCUGCUUA ((((.................----------............))))........((((((((.(((....)))((((.(......).))))...........))))))))......... ( -18.05) >consensus GGGAACCAAAACACCGCCCAC__________A_CACCCACACCUCCCACAGCCACCCACUGUCCCGAUAAUUCGUGUACAUCAUUUUUUACAGCCCCUCAUCAGUUGGUGGCUCUGCUUA ..............................................((.(((((((.((((...(((....)))((((.(......).)))).........)))).))))))).)).... (-15.10 = -16.30 + 1.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:14 2006