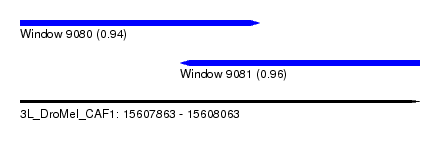
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,607,863 – 15,608,063 |
| Length | 200 |
| Max. P | 0.963553 |

| Location | 15,607,863 – 15,607,983 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -31.51 |
| Energy contribution | -31.76 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
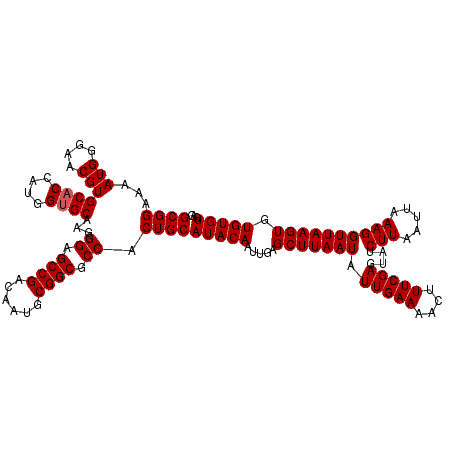
>3L_DroMel_CAF1 15607863 120 + 23771897 GCGGAAAAUGGGAACGUGCACCAUGGUGCAAGGGAGCCGACAAUGCGGCGCCACUGCAUACAAUUGAGCUUAAUAUUGAAAACUUUCGAGUAUCUUAAUUAAAGGUUAAGUGUGUGUGAU ((((...(((....)))((((....))))...((.((((......)))).)).))))(((((.....(((((((.(((((....)))))....(((.....)))))))))).)))))... ( -33.30) >DroSec_CAF1 31111 113 + 1 GCGGAAAAUGGGAACGUGCA-------GCAAGGGAGCCGACAAUGCGGCGCCACUGCAUACAAUUGAGCUUAAUAUUGAAAACUUUCGAGUAUCUUAAUUAAAGGUUAAGUGUGUGUGAG ((.(...(((....))).).-------)).((((.((((......)))).)).))(((((((.((((.(((...(((((..(((....)))...)))))..))).)))).)))))))... ( -29.10) >DroSim_CAF1 31126 120 + 1 GCGGAAAAUGGGAACGUGCACCAUGGUGCAAGGGAGCCGACAAUGCGGCGCCACUGCAUACAAUUGAGCUUAAUAUUGAAAACUUUCGAGUAUCUUAAUUAAAGGUUAAGUGUGUGUGAG ((((...(((....)))((((....))))...((.((((......)))).)).))))(((((.....(((((((.(((((....)))))....(((.....)))))))))).)))))... ( -33.30) >DroEre_CAF1 30996 120 + 1 GCGGAAAAUGGGAACGUGCACCAUGGUGCGAGGGCGCCGACAAUGCGGCGCCACUGCAUACAAUUGAGCUUAAUAUUGAAAACUUUCGAGUAUCUUAAUUAAAGGUUAAGUGUGUGUGAG ((((...(((....)))((((....))))...(((((((......))))))).))))(((((.....(((((((.(((((....)))))....(((.....)))))))))).)))))... ( -39.00) >DroYak_CAF1 33253 120 + 1 GCGGAAAAUGGGAACGUGCACCAUGGUGCAAGGGAGCCGACAAUGCGGCGCCACUGCAUACAAUUGAGCUUAAUAUUGAAAACUUUCGAGUAUCUUAAUUAAAGGUUAAGUGUGUGUGAG ((((...(((....)))((((....))))...((.((((......)))).)).))))(((((.....(((((((.(((((....)))))....(((.....)))))))))).)))))... ( -33.30) >consensus GCGGAAAAUGGGAACGUGCACCAUGGUGCAAGGGAGCCGACAAUGCGGCGCCACUGCAUACAAUUGAGCUUAAUAUUGAAAACUUUCGAGUAUCUUAAUUAAAGGUUAAGUGUGUGUGAG ((((...(((....)))((((....))))...((.((((......)))).)).))))(((((.....(((((((.(((((....)))))....(((.....)))))))))).)))))... (-31.51 = -31.76 + 0.25)

| Location | 15,607,943 – 15,608,063 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -16.63 |
| Consensus MFE | -15.44 |
| Energy contribution | -15.64 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
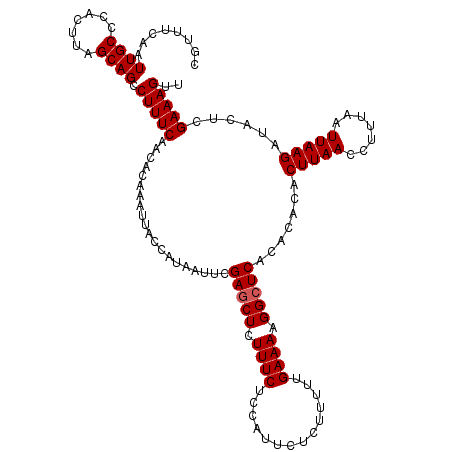
>3L_DroMel_CAF1 15607943 120 - 23771897 CGUUUCAAUUGCCCACUUAGCAGCCUUUCAACACAAAUUACCAUAAUUCGAGCUCUUUCUCCAUUCUCUUUUUGAAAAGGAUCACACACACUUAACCUUUAAUUAAGAUACUCGAAAGUU ........((((.......)))).......................((((((.......(((.((.((.....)).))))).........(((((.......)))))...)))))).... ( -11.80) >DroSec_CAF1 31184 120 - 1 CGUUUCAAUUGCCCACUUAGCAGCCUUUCAACACAAAUUACCAUAAUUCGAGCUCUUUCUCCAUUCUCUUUUUGAAAAGGCUCACACACACUUAACCUUUAAUUAAGAUACUCGAAAGUU ........((((.......)))).(((((....................((((..((((..............))))..)))).......(((((.......)))))......))))).. ( -17.14) >DroSim_CAF1 31206 120 - 1 CGUUUCAAUUGCCCACUUAGCAGCCUUUCAACACAAAUUACCAUAAUUCGAGCUCUUUCUCCAUUCUCUUUUUGAAAAGGCUCACACACACUUAACCUUUAAUUAAGAUACUCGAAAGUU ........((((.......)))).(((((....................((((..((((..............))))..)))).......(((((.......)))))......))))).. ( -17.14) >DroEre_CAF1 31076 120 - 1 CGUUUCAAUUGCCCACUUAGCAGCCUUUCAGCACAAAUUACCAUAAUUCGAGCUUUUUCUCCAUUCUCUUUUUGAAAAGGCUCACACACACUUAACCUUUAAUUAAGAUACUCGAAAGUU ........((((.......)))).(((((....................((((((((((..............)))))))))).......(((((.......)))))......))))).. ( -19.94) >DroYak_CAF1 33333 120 - 1 CGAUUCAAUUGCCCACUUAGCAGCCUUUCAACACAAAUUACCAUAAUUCGAGCUCUUUCUCCAUUCUCUUUUUGAAAAGGCUCACACACACUUAACCUUUAAUUAAGAUACUCGAAAGUU ........((((.......)))).(((((....................((((..((((..............))))..)))).......(((((.......)))))......))))).. ( -17.14) >consensus CGUUUCAAUUGCCCACUUAGCAGCCUUUCAACACAAAUUACCAUAAUUCGAGCUCUUUCUCCAUUCUCUUUUUGAAAAGGCUCACACACACUUAACCUUUAAUUAAGAUACUCGAAAGUU ........((((.......)))).(((((....................(((((.((((..............)))).))))).......(((((.......)))))......))))).. (-15.44 = -15.64 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:05 2006