
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,607,339 – 15,607,495 |
| Length | 156 |
| Max. P | 0.990955 |

| Location | 15,607,339 – 15,607,455 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.38 |
| Mean single sequence MFE | -30.72 |
| Consensus MFE | -28.50 |
| Energy contribution | -28.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15607339 116 + 23771897 AACCCCUUGCAGCCUCGACACCACCGCC----CAUCACAUGUGGCGCUUUUGCCUCAAUUAUUUUUAUUGGUUCGGCGCAAAAAUUUACGUUGCAUACUUUGCAGCGCCGCCUGGCAUGG ....((.(((.((.((((.((((..(((----(((...))).)))((....))...............)))))))).)).........(((((((.....))))))).......))).)) ( -31.50) >DroSec_CAF1 30589 119 + 1 AACCCCUUGCAGCCCCACCAACACCACC-CAUCAUCACAUGUGGCGCUUUUGCCUCAAUUAUUUUUAUUGGUUCGGCGCAAAAAUUUACGUUGCAUACUUUGCAGCGCCGCCUGGCAUGG ......((((.(((..(((((..((((.-...........)))).((....))..............)))))..)))))))........((((((.....))))))(((....))).... ( -30.00) >DroSim_CAF1 30601 120 + 1 AACCCCUUGCAGCCCCGCCAACACCAUCACAUCAUCACAUGUGGCGCUUUUGCCUCAAUUAUUUUUAUUGGUUCGGCGCAAAAAUUUACGUUGCAUACUUUGCAGCGCCGCCUGGCAUGG ......((((.(((..(((((.....((((((......)))))).((....))..............)))))..)))))))........((((((.....))))))(((....))).... ( -31.70) >DroEre_CAF1 30515 107 + 1 GCCCCCUU--AGCCCCGU------AACC----CAUCACAUGUGGCGCUUUUGCCUCAAUUAUUUUUAUUGGUUCGGCGCAAAAAUUUACGUUGCAUACUUUGCAGCGCCGCCU-GCAUGG ........--........------....----.....(((((((((.((((((((.(((((.......))))).)).)))))).....(((((((.....))))))).)))).-))))). ( -29.40) >DroYak_CAF1 32684 108 + 1 GCCCCCUU--AACCCCUU------AACC----CAUCACAUGUGGCGCUUUUGCCUCAAUUAUUUUUAUUGGUUCGGCGCAAAAAUUUACGUUGCAUACUUUGCAGCGCCGCCUGGCAUGG ........--........------...(----(((..((.(((((((((((((((.(((((.......))))).)).))))))........((((.....))))))))))).))..)))) ( -31.00) >consensus AACCCCUUGCAGCCCCGCCA_CACCACC____CAUCACAUGUGGCGCUUUUGCCUCAAUUAUUUUUAUUGGUUCGGCGCAAAAAUUUACGUUGCAUACUUUGCAGCGCCGCCUGGCAUGG .....................................(((((((((.((((((((.(((((.......))))).)).)))))).....(((((((.....))))))).))))..))))). (-28.50 = -28.50 + 0.00)

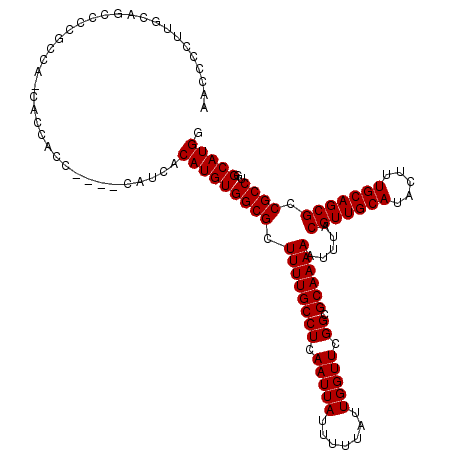

| Location | 15,607,375 – 15,607,495 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -30.06 |
| Consensus MFE | -30.10 |
| Energy contribution | -30.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15607375 120 - 23771897 ACAUGUCAAGUCGUUGCAAUUUUCCAUAACGAAAUUAACGCCAUGCCAGGCGGCGCUGCAAAGUAUGCAACGUAAAUUUUUGCGCCGAACCAAUAAAAAUAAUUGAGGCAAAAGCGCCAC ....((....((((((..........)))))).....))(((......)))((((((((((((..(((...)))...))))))(((....((((.......)))).)))...)))))).. ( -30.10) >DroSec_CAF1 30628 120 - 1 ACAUGUCAAGUCGUUGCAAUUUUCCAUAACGAAAUUAACGCCAUGCCAGGCGGCGCUGCAAAGUAUGCAACGUAAAUUUUUGCGCCGAACCAAUAAAAAUAAUUGAGGCAAAAGCGCCAC ....((....((((((..........)))))).....))(((......)))((((((((((((..(((...)))...))))))(((....((((.......)))).)))...)))))).. ( -30.10) >DroSim_CAF1 30641 120 - 1 ACAUGUCAAGUCGUUGCAAUUUUCCAUAACGAAAUUAACGCCAUGCCAGGCGGCGCUGCAAAGUAUGCAACGUAAAUUUUUGCGCCGAACCAAUAAAAAUAAUUGAGGCAAAAGCGCCAC ....((....((((((..........)))))).....))(((......)))((((((((((((..(((...)))...))))))(((....((((.......)))).)))...)))))).. ( -30.10) >DroEre_CAF1 30543 119 - 1 ACAUGUCAAGUCGUUGCAAUUUUCCAUAACGAAAUUAACGCCAUGC-AGGCGGCGCUGCAAAGUAUGCAACGUAAAUUUUUGCGCCGAACCAAUAAAAAUAAUUGAGGCAAAAGCGCCAC ....((....((((((..........)))))).....))(((....-.)))((((((((((((..(((...)))...))))))(((....((((.......)))).)))...)))))).. ( -29.90) >DroYak_CAF1 32712 120 - 1 ACAUGUCAAGUCGUUGCAAUUUUCCAUAACGAAAUUAACGCCAUGCCAGGCGGCGCUGCAAAGUAUGCAACGUAAAUUUUUGCGCCGAACCAAUAAAAAUAAUUGAGGCAAAAGCGCCAC ....((....((((((..........)))))).....))(((......)))((((((((((((..(((...)))...))))))(((....((((.......)))).)))...)))))).. ( -30.10) >consensus ACAUGUCAAGUCGUUGCAAUUUUCCAUAACGAAAUUAACGCCAUGCCAGGCGGCGCUGCAAAGUAUGCAACGUAAAUUUUUGCGCCGAACCAAUAAAAAUAAUUGAGGCAAAAGCGCCAC ....((....((((((..........)))))).....))(((......)))((((((((((((..(((...)))...))))))(((....((((.......)))).)))...)))))).. (-30.10 = -30.10 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:03 2006