| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,645,230 – 1,645,340 |
| Length | 110 |
| Max. P | 0.834902 |

| Location | 1,645,230 – 1,645,340 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.23 |
| Mean single sequence MFE | -41.17 |
| Consensus MFE | -20.88 |
| Energy contribution | -20.80 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.51 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
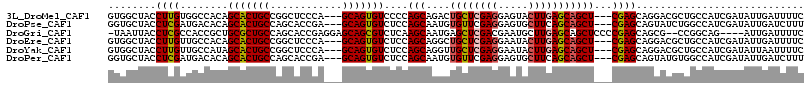
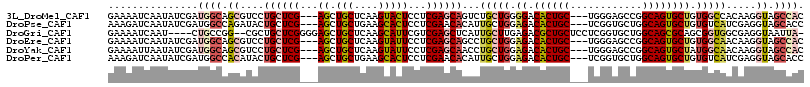
>3L_DroMel_CAF1 1645230 110 + 23771897 GUGGCUACCUUGUGGCCACAGCACUGCCGGCUCCCA---GCAGUGUCCCCAGCAGACUGCUCGAGGAGUACUUGAGCAGCU---CGAGCAGGACGCUGCCAUCGAUAUUGAUUUUC ((((((((...))))))))(((.......)))....---(((((((((...((.(((((((((((.....))))))))).)---)..)).)))))))))................. ( -47.80) >DroPse_CAF1 7935 110 + 1 GGUGCUACCUCGAUGACACAGCACUGCCAGCACCGA---GCAGUGUCUCCAGCAAUGUGUUCGAGGAGUGCUUCAGCAGCU---CGAGCAGUAUCUGGCCAUCGAUAUUGAUCUUU ((..((.((((((..(((((((((((((......).---))))))).........)))))))))))))..)).........---.((.(((((((........))))))).))... ( -36.40) >DroGri_CAF1 8735 109 + 1 -UAAUUACCUCGCCACCGCUGCGCUGCCAGCACCGAGGAGCAGCGUCUCAAGCAAUGAGCUCGACGAAUGCUUGAGCAGCUCCCCGAGCAGCG--CCGGCAG----AUUGAUUUUC -..........(((..((((((((.....))..((.(((((..((((...(((.....))).))))..(((....)))))))).)).))))))--..)))..----.......... ( -39.90) >DroEre_CAF1 7046 110 + 1 GUGGCUACCUUGUUGCCACAGCACUGCCGGCUCCCA---GCAGUGUCUCCAGCAGGCUGCUCGAGGAAUACUUGAGCAGCU---CGAGCAGGACGCUGCCAUCGAUAUUGAUUUUC (((((.........))))).(((((((.((...)).---))))))).........((((((((((.....))))))))))(---(((((((....))))..))))........... ( -45.60) >DroYak_CAF1 7222 110 + 1 GUGGCUACCUUGUUGCCAUAGCACUGCCGGCUCCCA---GCAGUGUCUCCAGCAGGUUGCUCGAGGAAUACUUGAGCAGCU---CGAGCAGGACGCUGCCAUCGAUAUUAAUUUUC (((((...((((((((...((((((((.((...)).---)))))).))...)).(((((((((((.....)))))))))))---..)))))).....))))).............. ( -41.00) >DroPer_CAF1 8015 110 + 1 GGUGCUACCUCGAUGACACAGCACUGCCAGCACCGA---GCAGUGUCUCCAGCAAUGUGUUCGAGGAGUGCUUCAGCAGCU---CGAGCAGUAUGUGGCCAUCGAUAUUGAUCUUU ((.....))((((((.((((((((((((......).---)))))))...........((((((((...(((....))).))---))))))...))))..))))))........... ( -36.30) >consensus GUGGCUACCUCGUUGCCACAGCACUGCCAGCACCCA___GCAGUGUCUCCAGCAAUGUGCUCGAGGAAUACUUGAGCAGCU___CGAGCAGGACGCUGCCAUCGAUAUUGAUUUUC ........((((........(((((((............)))))))....(((....((((((((.....)))))))))))...))))............................ (-20.88 = -20.80 + -0.08)
| Location | 1,645,230 – 1,645,340 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.23 |
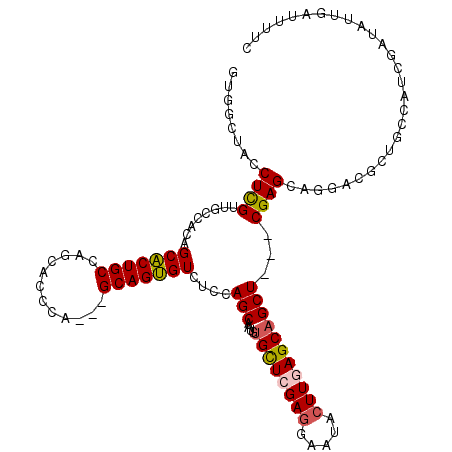
| Mean single sequence MFE | -44.44 |
| Consensus MFE | -21.02 |
| Energy contribution | -22.25 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
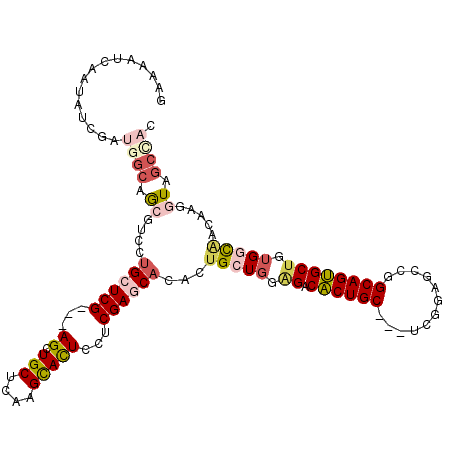
>3L_DroMel_CAF1 1645230 110 - 23771897 GAAAAUCAAUAUCGAUGGCAGCGUCCUGCUCG---AGCUGCUCAAGUACUCCUCGAGCAGUCUGCUGGGGACACUGC---UGGGAGCCGGCAGUGCUGUGGCCACAAGGUAGCCAC ....(((......)))..(((((..(((((((---((.(((....)))...)))))))))..))))).((.((((((---(((...)))))))))))(((((.((...)).))))) ( -46.90) >DroPse_CAF1 7935 110 - 1 AAAGAUCAAUAUCGAUGGCCAGAUACUGCUCG---AGCUGCUGAAGCACUCCUCGAACACAUUGCUGGAGACACUGC---UCGGUGCUGGCAGUGCUGUGUCAUCGAGGUAGCACC ...........(((((((((((.(((((((..---(((.(((((.(((((((.(((.....)))..))))....)))---)))))))))))))))))).))))))))......... ( -41.40) >DroGri_CAF1 8735 109 - 1 GAAAAUCAAU----CUGCCGG--CGCUGCUCGGGGAGCUGCUCAAGCAUUCGUCGAGCUCAUUGCUUGAGACGCUGCUCCUCGGUGCUGGCAGCGCAGCGGUGGCGAGGUAAUUA- ........((----((((((.--(((((((((((((((..((((((((..............)))))))).....))))))))).((.....)))))))).)))).)))).....- ( -50.74) >DroEre_CAF1 7046 110 - 1 GAAAAUCAAUAUCGAUGGCAGCGUCCUGCUCG---AGCUGCUCAAGUAUUCCUCGAGCAGCCUGCUGGAGACACUGC---UGGGAGCCGGCAGUGCUGUGGCAACAAGGUAGCCAC ...............((((.((...(((((((---((.(((....)))...)))))))))..(((((.((.((((((---(((...))))))))))).))))).....)).)))). ( -46.30) >DroYak_CAF1 7222 110 - 1 GAAAAUUAAUAUCGAUGGCAGCGUCCUGCUCG---AGCUGCUCAAGUAUUCCUCGAGCAACCUGCUGGAGACACUGC---UGGGAGCCGGCAGUGCUAUGGCAACAAGGUAGCCAC ...............((((.......((((((---((.(((....)))...))))))))((((((((.((.((((((---(((...))))))))))).))))....)))).)))). ( -42.10) >DroPer_CAF1 8015 110 - 1 AAAGAUCAAUAUCGAUGGCCACAUACUGCUCG---AGCUGCUGAAGCACUCCUCGAACACAUUGCUGGAGACACUGC---UCGGUGCUGGCAGUGCUGUGUCAUCGAGGUAGCACC ...........((((((((.((((((((((..---(((.(((((.(((((((.(((.....)))..))))....)))---))))))))))))))).)))))))))))......... ( -39.20) >consensus GAAAAUCAAUAUCGAUGGCAGCGUCCUGCUCG___AGCUGCUCAAGCACUCCUCGAGCACACUGCUGGAGACACUGC___UCGGAGCCGGCAGUGCUGUGGCAACAAGGUAGCCAC ...............((((.((....((((((...((.(((....)))))...))))))...(((((.((.((((((............)))))))).))))).....)).)))). (-21.02 = -22.25 + 1.23)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:39 2006