
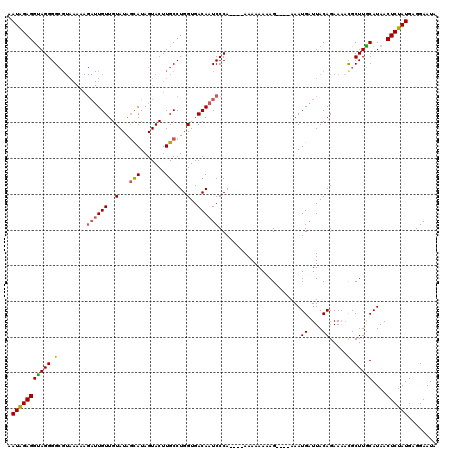
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,604,092 – 15,604,313 |
| Length | 221 |
| Max. P | 0.999711 |

| Location | 15,604,092 – 15,604,193 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.35 |
| Mean single sequence MFE | -22.68 |
| Consensus MFE | -16.71 |
| Energy contribution | -16.77 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15604092 101 - 23771897 GAUAGAGGUAGGGG-----------UGUUGUAUACUAUAGUACUUGCCUGGUGACAAUCCCA----AAAAAAAAG----AAAUGAUUACAGAAAACGCUUACAUAACUCUAUGAGGAAUA .(((((((((((.(-----------(.((((((((....)))......((....))......----.........----.......)))))...)).)))))....))))))........ ( -18.30) >DroSec_CAF1 27200 112 - 1 AAUAGAGGUAGGGGCGUAAAAAGAUUGUUGUAUAGCAUAGUACUUGCCUGGUGACAAUCCCA----AAAAAAUAG----AAAUGAUUACAGAAAACGCUUGCAUAACUCUAUGAGGAAUA .((((((.((.((((((.....((((((..(...(((.......)))...)..))))))...----......((.----...))..........))))))...)).))))))........ ( -23.60) >DroSim_CAF1 27114 111 - 1 GAUAGAGGUAGGGGCGUAAAAAGAUUGUUGUAUAGCAUAGUACUUGCCUGGUGACAAUCCCA----AAAAAAUAG----AAAUGAUUACAGAA-AUGCUUGCAUAACUCUAUGAGGAAUA .((((((.((.((((((.....((((((..(...(((.......)))...)..))))))...----......((.----...)).........-))))))...)).))))))........ ( -21.90) >DroYak_CAF1 29226 120 - 1 AAUGGAGGCAGGGGCGUAAAAAGAUUGUUGUAUGGCAUAGUACUUGCCUGGUGACAAUCCCAAUACAAAAAAAAGAAAAAAAUGAUUACAGAAACCGCUUGCAUAACUCUAUGAGGAAUU .(((((((((((((.(((....((((((..(..((((.......))))..)..))))))....)))................((....))....)).)))))....))))))........ ( -26.90) >consensus AAUAGAGGUAGGGGCGUAAAAAGAUUGUUGUAUAGCAUAGUACUUGCCUGGUGACAAUCCCA____AAAAAAAAG____AAAUGAUUACAGAAAACGCUUGCAUAACUCUAUGAGGAAUA .(((((((((((.(........((((((..(...(((.......)))...)..)))))).......................((....)).....).)))))....))))))........ (-16.71 = -16.77 + 0.06)


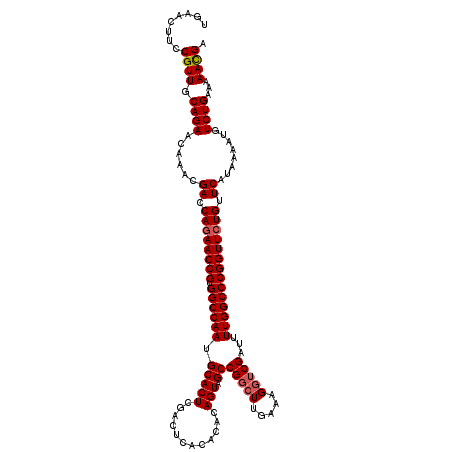
| Location | 15,604,193 – 15,604,313 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -41.01 |
| Consensus MFE | -35.88 |
| Energy contribution | -36.24 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.46 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.93 |
| SVM RNA-class probability | 0.999711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15604193 120 + 23771897 UGAACUUCCGUUGCAGAACAAACGACCAGAACCGUGGCCAAUGCACUCGACACACACACAGUAGCCGGCUUGAAAGGUCGAUUUUGGCCCGGUUCUGUUCAUAAAAAGUCUGAAAAACGA ........((((.((((......((.((((((((.((((((.....(((((.....((.(((.....)))))....)))))..)))))))))))))).))........))))...)))). ( -40.04) >DroSec_CAF1 27312 120 + 1 UGAACUUCCGUUGCAGAACAAACGACCAGAACCGUGGCCAAUGCACUGGACUCACACACAGUAGCCGGCUUGAAAGGUCGAUUUUGGCCCGGUUCUGUUCAUAAAAUGUCUGAAAAACGA ........((((.((((......((.((((((((.((((((.((((((..........)))).))(((((.....)))))...)))))))))))))).))........))))...)))). ( -43.44) >DroSim_CAF1 27225 120 + 1 UGAACUUCCGUUGCAGAACAAACGACCAGAACCGUGGCCAAUGCACUCGACUCACACACAGUAGCCGGCUUGAAAGGUCGAUAUUGGCCCGGUUCUGUUCAUAAAAUGUCUGAAAAACGA ........((((.((((......((.((((((((.((((((((...((((((....((.(((.....)))))...)))))))))))))))))))))).))........))))...)))). ( -42.84) >DroEre_CAF1 27211 120 + 1 UAAACUUCCGUUGCAGAACAAACGACCAGAACCGUGGCCAAUGCACUCGACUCACUCACAGUAGCCGGCUUGAAAGGUCGAUUUUGGCCCGGUUGUGUUCGUAAAAUGUCUGAAAAAUGA ........((((.((((....((((.((.(((((.((((((.....((((((...(((.(((.....))))))..))))))..))))))))))).)).))))......))))...)))). ( -41.00) >DroYak_CAF1 29346 120 + 1 CGAACUUCCGUUGCAGAACAAACGACCAGAACCGUGGCCAAUGCACUCGACUCACACACAGUAGCCGCAUUGAAAGGUCGAUUUUGGCCCGGUUCUGUUCAUAAAAUGUCUGAAAAAUGA ........((((.((((......((.((((((((.((((((.....((((((......((((......))))...))))))..)))))))))))))).))........))))...)))). ( -37.74) >consensus UGAACUUCCGUUGCAGAACAAACGACCAGAACCGUGGCCAAUGCACUCGACUCACACACAGUAGCCGGCUUGAAAGGUCGAUUUUGGCCCGGUUCUGUUCAUAAAAUGUCUGAAAAACGA ........((((.((((......((.((((((((.((((((.(((((............))).))(((((.....)))))...)))))))))))))).))........))))...)))). (-35.88 = -36.24 + 0.36)

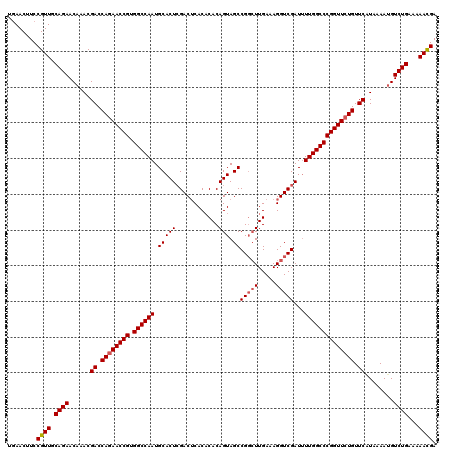
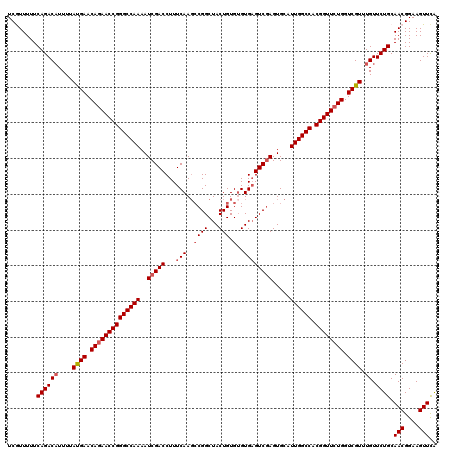
| Location | 15,604,193 – 15,604,313 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -41.82 |
| Consensus MFE | -38.18 |
| Energy contribution | -39.02 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.88 |
| SVM RNA-class probability | 0.999679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15604193 120 - 23771897 UCGUUUUUCAGACUUUUUAUGAACAGAACCGGGCCAAAAUCGACCUUUCAAGCCGGCUACUGUGUGUGUGUCGAGUGCAUUGGCCACGGUUCUGGUCGUUUGUUCUGCAACGGAAGUUCA (((((...((((......((((.((((((((((((((..(((((....((.(((((...))).)).)).))))).....)))))).)))))))).))))....)))).)))))....... ( -41.30) >DroSec_CAF1 27312 120 - 1 UCGUUUUUCAGACAUUUUAUGAACAGAACCGGGCCAAAAUCGACCUUUCAAGCCGGCUACUGUGUGUGAGUCCAGUGCAUUGGCCACGGUUCUGGUCGUUUGUUCUGCAACGGAAGUUCA (((((...((((((....((((.((((((((((((((....((....))......((.((((..(....)..)))))).)))))).)))))))).)))).)).)))).)))))....... ( -40.20) >DroSim_CAF1 27225 120 - 1 UCGUUUUUCAGACAUUUUAUGAACAGAACCGGGCCAAUAUCGACCUUUCAAGCCGGCUACUGUGUGUGAGUCGAGUGCAUUGGCCACGGUUCUGGUCGUUUGUUCUGCAACGGAAGUUCA (((((...((((((....((((.(((((((((((((((.(((((...(((.(((((...))).)).))))))))....))))))).)))))))).)))).)).)))).)))))....... ( -45.50) >DroEre_CAF1 27211 120 - 1 UCAUUUUUCAGACAUUUUACGAACACAACCGGGCCAAAAUCGACCUUUCAAGCCGGCUACUGUGAGUGAGUCGAGUGCAUUGGCCACGGUUCUGGUCGUUUGUUCUGCAACGGAAGUUUA ........((((((....((((.((.(((((((((((..(((((...(((..((((...))).)..)))))))).....)))))).))))).)).)))).)).)))).(((....))).. ( -38.50) >DroYak_CAF1 29346 120 - 1 UCAUUUUUCAGACAUUUUAUGAACAGAACCGGGCCAAAAUCGACCUUUCAAUGCGGCUACUGUGUGUGAGUCGAGUGCAUUGGCCACGGUUCUGGUCGUUUGUUCUGCAACGGAAGUUCG ........((((((....((((.((((((((((((((..(((((...(((((((((...)))))).)))))))).....)))))).)))))))).)))).)).)))).(((....))).. ( -43.60) >consensus UCGUUUUUCAGACAUUUUAUGAACAGAACCGGGCCAAAAUCGACCUUUCAAGCCGGCUACUGUGUGUGAGUCGAGUGCAUUGGCCACGGUUCUGGUCGUUUGUUCUGCAACGGAAGUUCA ........((((((....((((.((((((((((((((..(((((...(((..((((...))).)..)))))))).....)))))).)))))))).)))).)).)))).(((....))).. (-38.18 = -39.02 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:56 2006