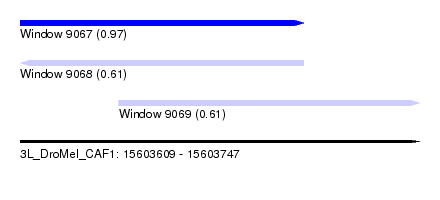
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,603,609 – 15,603,747 |
| Length | 138 |
| Max. P | 0.965930 |

| Location | 15,603,609 – 15,603,707 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.11 |
| Mean single sequence MFE | -15.32 |
| Consensus MFE | -13.32 |
| Energy contribution | -13.04 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15603609 98 + 23771897 AUCUUUUAUCCAAACACAACAAGCCUCAU------UCUUAACC----------------UCCGACCCUUCGAACAAAUGCCGUCAGUUGUUCGAACACUCCUCUUUGUGAGAUAUCUCCC ......((((...................------........----------------........(((((((((.((....)).)))))))))(((........))).))))...... ( -13.40) >DroSec_CAF1 26744 94 + 1 AUCUUUUAUCCAAACACAACAAGCCUCAU------UCUCAACC----------------UCCGACCCUUCGAACAAAUGCCGUCAGUUGUUCGAACACUCCUCUUUGUGGGAUAUC---- ......(((((..................------........----------------........(((((((((.((....)).)))))))))(((........))))))))..---- ( -17.60) >DroSim_CAF1 26652 100 + 1 AUCUUUUAUCCAAACACAACAAGCCUCAUUCUCAUUCUCAAUC----------------UCCGACCCUUCGACCAAAUGCCGUCAGUUGUUCGAACACUCCUCCUUGUGGGAUAUC---- ......(((((....(((((.......................----------------...((....))(((........))).))))).....(((........))))))))..---- ( -13.50) >DroEre_CAF1 26709 92 + 1 AUCUUUUAUCCAAACACAACAAGCCUCAA------UCUCAACC----------------UCCGACCCUUCGAGCAAAUGCCGUCAGUUGCUCGAACACUCCUCUUUGUGAGAUA------ ............................(------(((((...----------------...((...(((((((((.((....)).)))))))))...)).......)))))).------ ( -18.62) >DroYak_CAF1 28734 110 + 1 AUCUUUUAUCCAAACACAACAAGCCUCAU------UCUCAACCUCAGUACCUCAGACCCUCCGACCCUUCGAACAAAUGCCGUCAGUUGUUCGAACACUCCUCUUUGUGAGAUAUC---- ......((((....(((((..((......------................((.(.....).))...(((((((((.((....)).))))))))).....))..))))).))))..---- ( -13.50) >consensus AUCUUUUAUCCAAACACAACAAGCCUCAU______UCUCAACC________________UCCGACCCUUCGAACAAAUGCCGUCAGUUGUUCGAACACUCCUCUUUGUGAGAUAUC____ ((((...............................................................(((((((((.((....)).)))))))))(((........)))))))....... (-13.32 = -13.04 + -0.28)


| Location | 15,603,609 – 15,603,707 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.11 |
| Mean single sequence MFE | -25.14 |
| Consensus MFE | -21.86 |
| Energy contribution | -21.58 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15603609 98 - 23771897 GGGAGAUAUCUCACAAAGAGGAGUGUUCGAACAACUGACGGCAUUUGUUCGAAGGGUCGGA----------------GGUUAAGA------AUGAGGCUUGUUGUGUUUGGAUAAAAGAU ......((((((((((....((.(.(((((((((.((....)).))))))))).).))..(----------------((((....------....))))).)))))...)))))...... ( -23.40) >DroSec_CAF1 26744 94 - 1 ----GAUAUCCCACAAAGAGGAGUGUUCGAACAACUGACGGCAUUUGUUCGAAGGGUCGGA----------------GGUUGAGA------AUGAGGCUUGUUGUGUUUGGAUAAAAGAU ----..((((((((((.(((...(((((...(((((..((((.(((....)))..))))..----------------))))).))------)))...))).)))))...)))))...... ( -26.90) >DroSim_CAF1 26652 100 - 1 ----GAUAUCCCACAAGGAGGAGUGUUCGAACAACUGACGGCAUUUGGUCGAAGGGUCGGA----------------GAUUGAGAAUGAGAAUGAGGCUUGUUGUGUUUGGAUAAAAGAU ----..((((((((((.(((.(((.(((((....((..((((.....))))..)).)))))----------------.)))....((....))....))).)))))...)))))...... ( -22.90) >DroEre_CAF1 26709 92 - 1 ------UAUCUCACAAAGAGGAGUGUUCGAGCAACUGACGGCAUUUGCUCGAAGGGUCGGA----------------GGUUGAGA------UUGAGGCUUGUUGUGUUUGGAUAAAAGAU ------((((((((((....((.(.(((((((((.((....)).))))))))).).))..(----------------((((....------....))))).)))))...)))))...... ( -25.80) >DroYak_CAF1 28734 110 - 1 ----GAUAUCUCACAAAGAGGAGUGUUCGAACAACUGACGGCAUUUGUUCGAAGGGUCGGAGGGUCUGAGGUACUGAGGUUGAGA------AUGAGGCUUGUUGUGUUUGGAUAAAAGAU ----..((((((((((....((.(.(((((((((.((....)).))))))))).).))...((((((.(...((....)).....------.).)))))).)))))...)))))...... ( -26.70) >consensus ____GAUAUCUCACAAAGAGGAGUGUUCGAACAACUGACGGCAUUUGUUCGAAGGGUCGGA________________GGUUGAGA______AUGAGGCUUGUUGUGUUUGGAUAAAAGAU ......((((((((((.(((((.(.(((((((((.((....)).))))))))).).)).....................((............))..))).)))))...)))))...... (-21.86 = -21.58 + -0.28)



| Location | 15,603,643 – 15,603,747 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.01 |
| Mean single sequence MFE | -19.88 |
| Consensus MFE | -15.71 |
| Energy contribution | -16.27 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15603643 104 + 23771897 ACC----------------UCCGACCCUUCGAACAAAUGCCGUCAGUUGUUCGAACACUCCUCUUUGUGAGAUAUCUCCCUAUCUCCCGAUGUUCUCGGUUCCCGAUUCCCCGUUUUGCC ...----------------.((((...(((((((((.((....)).))))))))).........(((.((((((......)))))).))).....))))..................... ( -20.60) >DroSec_CAF1 26778 86 + 1 ACC----------------UCCGACCCUUCGAACAAAUGCCGUCAGUUGUUCGAACACUCCUCUUUGUGGGAUAUC--------UCCCGAUGUUCCCGGUCCCCGAUUCC---------- ...----------------...((((.(((((((((.((....)).)))))))))...........(.((((((((--------....))))))))))))).........---------- ( -24.10) >DroSim_CAF1 26692 86 + 1 AUC----------------UCCGACCCUUCGACCAAAUGCCGUCAGUUGUUCGAACACUCCUCCUUGUGGGAUAUC--------UCCCGAUGUUCCCGGUCCCCGAUUCC---------- ...----------------...((((.(((((.(((.((....)).))).)))))...........(.((((((((--------....))))))))))))).........---------- ( -19.90) >DroYak_CAF1 28768 109 + 1 ACCUCAGUACCUCAGACCCUCCGACCCUUCGAACAAAUGCCGUCAGUUGUUCGAACACUCCUCUUUGUGAGAUAUC--------UCCCCAUGUUCCCGGCCCCCGAUCCCCCGAUAU--- ....................(((....(((((((((.((....)).)))))))))(((........))).......--------............)))..................--- ( -14.90) >consensus ACC________________UCCGACCCUUCGAACAAAUGCCGUCAGUUGUUCGAACACUCCUCUUUGUGAGAUAUC________UCCCGAUGUUCCCGGUCCCCGAUUCC__________ ......................((((.(((((((((.((....)).))))))))).............((((((((............)))).))))))))................... (-15.71 = -16.27 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:53 2006