
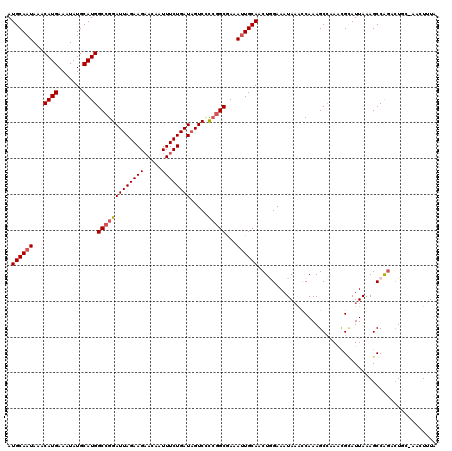
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,600,829 – 15,601,027 |
| Length | 198 |
| Max. P | 0.997669 |

| Location | 15,600,829 – 15,600,947 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.29 |
| Mean single sequence MFE | -26.41 |
| Consensus MFE | -18.28 |
| Energy contribution | -18.72 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15600829 118 - 23771897 AUGCAAUAAACAUGAAAUAUGCAUGGCCGGAUUAGAAGAACAAUUUCUGAUUGUCCCCGGCGAAAUUGCAACUGGAAAUAAAUCAAAGCCAAACGCAUUAAAGCCAGAAUGC--ACUUUA .((((((...((((.......))))(((((((((((((.....)))))))).....)))))...))))))..(((.............)))...(((((........)))))--...... ( -26.62) >DroSec_CAF1 23889 119 - 1 AUGCAAUAAACAUGAAAUAUGCAUGGCCGGAUUAGAAGAACAAUUUCUGAUAGUCGCCGGCGAAAUUGCAACUGGAAAUAAACCAAAGCCAAAUGAAUUAAAGCCAUACUAA-AACUUUU .((((((...((((.......))))(((((((((..((((....))))..))))..)))))...))))))..(((...(((..((........))..)))...)))......-....... ( -23.40) >DroSim_CAF1 23803 118 - 1 AUGCAAUAAACAUGAAAUAUGCAUGGCCGGAUUAGAAGAACAAUUUCUGAUAGUCCUCUGCGAAAUUGCAACUGGAAAUAAACCAAAGCCAAAUGCAUUAAAGCCAGACUGC--ACUUUA .((((.....((((.......))))...((((((..((((....))))..))))))(((((.....((((..(((.............)))..)))).....).)))).)))--)..... ( -25.62) >DroEre_CAF1 24005 119 - 1 AUGCAAUAAACAUGAAAUAUGCAUGGCCGGAUUAGAAGAACAAUUUCUGAUAGUC-CGGGCAAAAUUGCAACUAGAAAUAAGCCAAAGCGAAGCGUAUUAAAGCCGGACUGGUAAAUAUA .((((((...((((.......)))).((((((((..((((....))))..)))))-))).....))))))(((((......((....))...((........))....)))))....... ( -30.90) >DroYak_CAF1 25510 120 - 1 AUGCAAUAAACAUGAAAUAUGCAUGGCCGGAUUAGAAGCACAAUUUCUGAUAGUCUCCGGCAAAAGUGCAACUAGAAAUAUACCAAAGCCGAACGCAUUAAAGCCAGACUGGUAGAUAUU ((((...............(((((.(((((((((((((.....))))))).....))))))....)))))..((....))..............))))....(((.....)))....... ( -25.50) >consensus AUGCAAUAAACAUGAAAUAUGCAUGGCCGGAUUAGAAGAACAAUUUCUGAUAGUCCCCGGCGAAAUUGCAACUGGAAAUAAACCAAAGCCAAACGCAUUAAAGCCAGACUGC_AACUUUA .((((((...((((.......))))(((((((((((((.....)))))))).....)))))...)))))).................................................. (-18.28 = -18.72 + 0.44)


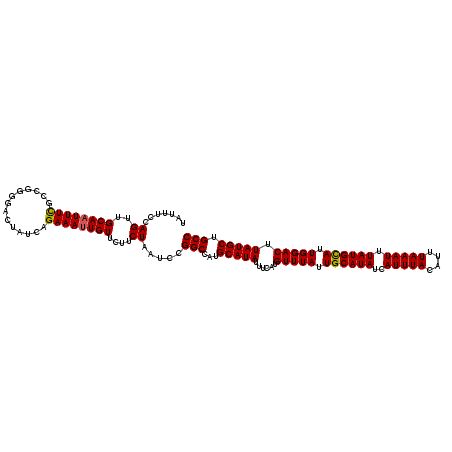
| Location | 15,600,867 – 15,600,987 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.92 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -22.96 |
| Energy contribution | -22.76 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15600867 120 + 23771897 UAUUUCCAGUUGCAAUUUCGCCGGGGACAAUCAGAAAUUGUUCUUCUAAUCCGGCCAUGCAUAUUUCAUGUUUAUUGCAUAUCAUUUACAUUUAAAUUUAUGUAUUGGACUUAUGCUGCC ...........(((.....((((((((((((.....))))))).......)))))...(((((......(((((.((((((..(((((....))))).)))))).))))).)))))))). ( -27.21) >DroSec_CAF1 23928 120 + 1 UAUUUCCAGUUGCAAUUUCGCCGGCGACUAUCAGAAAUUGUUCUUCUAAUCCGGCCAUGCAUAUUUCAUGUUUAUUGCAUAUCAUUUACAUUUAAAUUUAUGCAUUGGACUUAUGCUGCC ...........(((.....(((((........((((....))))......)))))...(((((......(((((.((((((..(((((....))))).)))))).))))).)))))))). ( -25.64) >DroSim_CAF1 23841 120 + 1 UAUUUCCAGUUGCAAUUUCGCAGAGGACUAUCAGAAAUUGUUCUUCUAAUCCGGCCAUGCAUAUUUCAUGUUUAUUGCAUAUCAUUUACAUUUAAAUUUAUGCAUUGGACUUAUGCUGCC .......((..((((((((...((......)).))))))))....)).....(((...(((((......(((((.((((((..(((((....))))).)))))).))))).))))).))) ( -25.90) >DroEre_CAF1 24045 119 + 1 UAUUUCUAGUUGCAAUUUUGCCCG-GACUAUCAGAAAUUGUUCUUCUAAUCCGGCCAUGCAUAUUUCAUGUUUAUUGCAUAUCAUUUACAUUUAAAUUUAUGCAUUGGACUUAUGCUGCC ...........(((....((.(((-((.((..((((....))))..)).))))).)).(((((......(((((.((((((..(((((....))))).)))))).))))).)))))))). ( -27.20) >DroYak_CAF1 25550 120 + 1 UAUUUCUAGUUGCACUUUUGCCGGAGACUAUCAGAAAUUGUGCUUCUAAUCCGGCCAUGCAUAUUUCAUGUUUAUUGCAUAUCAUUUACAUUUAAAUUUAUGCAUUGGACUUAUGCUGCC ......(((..(((((((((..(....)...)))))...))))..)))....(((...(((((......(((((.((((((..(((((....))))).)))))).))))).))))).))) ( -27.90) >consensus UAUUUCCAGUUGCAAUUUCGCCGGGGACUAUCAGAAAUUGUUCUUCUAAUCCGGCCAUGCAUAUUUCAUGUUUAUUGCAUAUCAUUUACAUUUAAAUUUAUGCAUUGGACUUAUGCUGCC .......((..((((((((..............))))))))....)).....(((...(((((......(((((.((((((..(((((....))))).)))))).))))).))))).))) (-22.96 = -22.76 + -0.20)


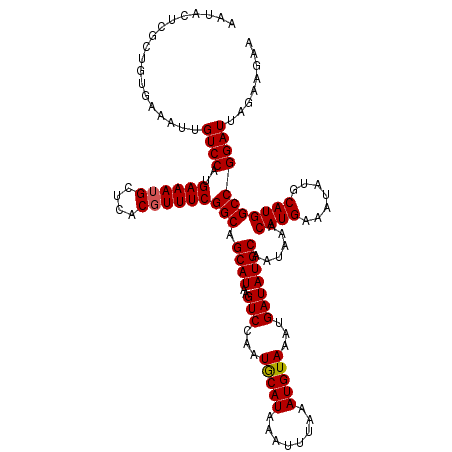
| Location | 15,600,867 – 15,600,987 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.92 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -23.38 |
| Energy contribution | -23.82 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15600867 120 - 23771897 GGCAGCAUAAGUCCAAUACAUAAAUUUAAAUGUAAAUGAUAUGCAAUAAACAUGAAAUAUGCAUGGCCGGAUUAGAAGAACAAUUUCUGAUUGUCCCCGGCGAAAUUGCAACUGGAAAUA ...........((((.(((((........))))).......((((((...((((.......))))(((((((((((((.....)))))))).....)))))...))))))..)))).... ( -28.10) >DroSec_CAF1 23928 120 - 1 GGCAGCAUAAGUCCAAUGCAUAAAUUUAAAUGUAAAUGAUAUGCAAUAAACAUGAAAUAUGCAUGGCCGGAUUAGAAGAACAAUUUCUGAUAGUCGCCGGCGAAAUUGCAACUGGAAAUA ...........((((.(((((........))))).......((((((...((((.......))))(((((((((..((((....))))..))))..)))))...))))))..)))).... ( -28.50) >DroSim_CAF1 23841 120 - 1 GGCAGCAUAAGUCCAAUGCAUAAAUUUAAAUGUAAAUGAUAUGCAAUAAACAUGAAAUAUGCAUGGCCGGAUUAGAAGAACAAUUUCUGAUAGUCCUCUGCGAAAUUGCAACUGGAAAUA (((.(((((..((...((((((.(((((....)))))..))))))........))..)))))...)))((((((..((((....))))..))))))..((((....)))).......... ( -28.20) >DroEre_CAF1 24045 119 - 1 GGCAGCAUAAGUCCAAUGCAUAAAUUUAAAUGUAAAUGAUAUGCAAUAAACAUGAAAUAUGCAUGGCCGGAUUAGAAGAACAAUUUCUGAUAGUC-CGGGCAAAAUUGCAACUAGAAAUA .((((((((..((...((((((.(((((....)))))..))))))........))..))))).((.((((((((..((((....))))..)))))-))).))....)))........... ( -33.40) >DroYak_CAF1 25550 120 - 1 GGCAGCAUAAGUCCAAUGCAUAAAUUUAAAUGUAAAUGAUAUGCAAUAAACAUGAAAUAUGCAUGGCCGGAUUAGAAGCACAAUUUCUGAUAGUCUCCGGCAAAAGUGCAACUAGAAAUA .((((((((..((...((((((.(((((....)))))..))))))........))..)))))...(((((((((((((.....))))))).....)))))).....)))........... ( -28.70) >consensus GGCAGCAUAAGUCCAAUGCAUAAAUUUAAAUGUAAAUGAUAUGCAAUAAACAUGAAAUAUGCAUGGCCGGAUUAGAAGAACAAUUUCUGAUAGUCCCCGGCGAAAUUGCAACUGGAAAUA .((((((((..((...((((((.(((((....)))))..))))))........))..)))))...(((((((((((((.....)))))))).....))))).....)))........... (-23.38 = -23.82 + 0.44)

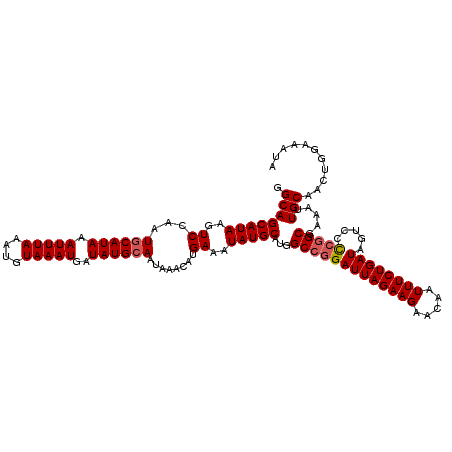

| Location | 15,600,907 – 15,601,027 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -24.70 |
| Energy contribution | -24.54 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15600907 120 + 23771897 UUCUUCUAAUCCGGCCAUGCAUAUUUCAUGUUUAUUGCAUAUCAUUUACAUUUAAAUUUAUGUAUUGGACUUAUGCUGCCGAAACGUGAGCAUUUCAUGGACAAUUUCACAGCGAGUAUU ............(.(((((.....(((((((((...(((((((...(((((........)))))...))..))))).....))))))))).....))))).).................. ( -24.20) >DroSec_CAF1 23968 120 + 1 UUCUUCUAAUCCGGCCAUGCAUAUUUCAUGUUUAUUGCAUAUCAUUUACAUUUAAAUUUAUGCAUUGGACUUAUGCUGCCGAAACGUGAGCAUUUCAUGGACAAUUUCACAGCGAGUAUU .........((((((...(((((......(((((.((((((..(((((....))))).)))))).))))).))))).)))(((((....)..))))..)))................... ( -25.60) >DroSim_CAF1 23881 117 + 1 UUCUUCUAAUCCGGCCAUGCAUAUUUCAUGUUUAUUGCAUAUCAUUUACAUUUAAAUUUAUGCAUUGGACUUAUGCUGCCGAAA-GGGAGCAUUUCAUGGACAAU-UCACAG-GAAUAUU (((((.........(((((..........(((((.((((((..(((((....))))).)))))).)))))..(((((.((....-)).)))))..))))).....-....))-))).... ( -30.87) >DroEre_CAF1 24084 120 + 1 UUCUUCUAAUCCGGCCAUGCAUAUUUCAUGUUUAUUGCAUAUCAUUUACAUUUAAAUUUAUGCAUUGGACUUAUGCUGCCGAAACGUGAGCAUUUCAUGGACAAUUUCACAGCGAGUAUU .........((((((...(((((......(((((.((((((..(((((....))))).)))))).))))).))))).)))(((((....)..))))..)))................... ( -25.60) >DroYak_CAF1 25590 120 + 1 UGCUUCUAAUCCGGCCAUGCAUAUUUCAUGUUUAUUGCAUAUCAUUUACAUUUAAAUUUAUGCAUUGGACUUAUGCUGCCGAAACGUGAGCAUUUCAUGGACAAUUUCACAGCGAGUAUU (((((((....((((...(((((......(((((.((((((..(((((....))))).)))))).))))).))))).))))...((((((...))))))...........)).))))).. ( -28.00) >consensus UUCUUCUAAUCCGGCCAUGCAUAUUUCAUGUUUAUUGCAUAUCAUUUACAUUUAAAUUUAUGCAUUGGACUUAUGCUGCCGAAACGUGAGCAUUUCAUGGACAAUUUCACAGCGAGUAUU .........((((((...(((((......(((((.((((((..(((((....))))).)))))).))))).))))).)))((((.(....).))))..)))................... (-24.70 = -24.54 + -0.16)



| Location | 15,600,907 – 15,601,027 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -25.84 |
| Consensus MFE | -23.52 |
| Energy contribution | -23.56 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15600907 120 - 23771897 AAUACUCGCUGUGAAAUUGUCCAUGAAAUGCUCACGUUUCGGCAGCAUAAGUCCAAUACAUAAAUUUAAAUGUAAAUGAUAUGCAAUAAACAUGAAAUAUGCAUGGCCGGAUUAGAAGAA ..(((.....))).....((((..((((((....))))))(((.((((..(((...(((((........)))))...)))))))......((((.......)))))))))))........ ( -24.40) >DroSec_CAF1 23968 120 - 1 AAUACUCGCUGUGAAAUUGUCCAUGAAAUGCUCACGUUUCGGCAGCAUAAGUCCAAUGCAUAAAUUUAAAUGUAAAUGAUAUGCAAUAAACAUGAAAUAUGCAUGGCCGGAUUAGAAGAA .....((((((((((((((..(((...)))..)).))))).)))))...(((((..((((((.(((((....)))))..)))))).....((((.......))))...))))).)).... ( -26.20) >DroSim_CAF1 23881 117 - 1 AAUAUUC-CUGUGA-AUUGUCCAUGAAAUGCUCCC-UUUCGGCAGCAUAAGUCCAAUGCAUAAAUUUAAAUGUAAAUGAUAUGCAAUAAACAUGAAAUAUGCAUGGCCGGAUUAGAAGAA ....(((-.(.(((-.(((.(((((..(((((.((-....)).)))))........((((((.(((((....)))))..))))))................))))).))).))).).))) ( -24.00) >DroEre_CAF1 24084 120 - 1 AAUACUCGCUGUGAAAUUGUCCAUGAAAUGCUCACGUUUCGGCAGCAUAAGUCCAAUGCAUAAAUUUAAAUGUAAAUGAUAUGCAAUAAACAUGAAAUAUGCAUGGCCGGAUUAGAAGAA .....((((((((((((((..(((...)))..)).))))).)))))...(((((..((((((.(((((....)))))..)))))).....((((.......))))...))))).)).... ( -26.20) >DroYak_CAF1 25590 120 - 1 AAUACUCGCUGUGAAAUUGUCCAUGAAAUGCUCACGUUUCGGCAGCAUAAGUCCAAUGCAUAAAUUUAAAUGUAAAUGAUAUGCAAUAAACAUGAAAUAUGCAUGGCCGGAUUAGAAGCA .......(((........((((..((((((....))))))(((.(((((..((...((((((.(((((....)))))..))))))........))..)))))...)))))))....))). ( -28.40) >consensus AAUACUCGCUGUGAAAUUGUCCAUGAAAUGCUCACGUUUCGGCAGCAUAAGUCCAAUGCAUAAAUUUAAAUGUAAAUGAUAUGCAAUAAACAUGAAAUAUGCAUGGCCGGAUUAGAAGAA ..................((((..((((((....))))))(((.((((..(((...(((((........)))))...)))))))......((((.......)))))))))))........ (-23.52 = -23.56 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:48 2006