| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,642,393 – 1,642,495 |
| Length | 102 |
| Max. P | 0.708477 |

| Location | 1,642,393 – 1,642,495 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.66 |
| Mean single sequence MFE | -18.74 |
| Consensus MFE | -14.53 |
| Energy contribution | -14.87 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
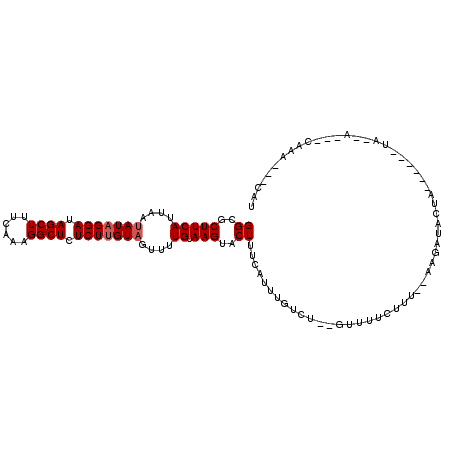
>3L_DroMel_CAF1 1642393 102 - 23771897 GGCGCUUCAUUAAUAUAGGAUAGCUUUCAAUGGCUCUCUUGUAGUUUUGUAAGUACUUUCAUUUGACU--GUUCUCUUU--AAGCUACUUAUUGUAUU--A---CAAC---CAU ((..............((((.((((......)))).))))(((((...((((((((((......((..--..)).....--))).)))))))...)))--)---)..)---).. ( -18.50) >DroVir_CAF1 4881 104 - 1 GGCGCUUCAUUGAUAUAGGAUAGCUUUCAAAGGCUCUCUUGUAGUUUUGUAAGUACUUUCAUUUGUCU--GUUUUCUUU--AACAUAUUA------UACAUAUAUAUAUAGCAU .((((((((....(((((((.(((((....))))).)))))))....)).)))).............(--(((......--)))).....------..............)).. ( -18.00) >DroGri_CAF1 5062 100 - 1 GGCGCUUCAUUGAUAUAGGAUAGCUUUCAAAGGCUCUCUUGUAGUUUUGUAAGUACUUUCAUUUGUCUCUGUUUUCUUU--AAGAUAUUA------UAUAG---CAUA---UAU .((((((((....(((((((.(((((....))))).)))))))....)).)))).........(((((..(......).--.)))))...------....)---)...---... ( -18.60) >DroMoj_CAF1 5095 100 - 1 GGCGCUUCAUUAAUAUAGGAUAGCUUUCAAAGGCUCUCUUGUAGUUUUGUAAGUACUUUCAUUUGUCU--GUUUUCUUUCUAACAUAU---------AAAUAUAUAUU---UCU ((..(((((....(((((((.(((((....))))).)))))))....)).)))..))...((((((.(--(((........)))).))---------)))).......---... ( -19.90) >DroAna_CAF1 4198 97 - 1 GGCGCUUCAUUAAAAUAGGAUAGCUUUCAAUGGCUCUCUAGUAGUUUUGUAAGUACU-UCAUUU------GUUUUAUUU--AAGCUACCAAUCGCCGC--A---GAAC---UCU ((((.............(((.((((......)))).))).(((((((.(((((.((.-......------)))))))..--)))))))....))))..--.---....---... ( -20.90) >DroPer_CAF1 4583 102 - 1 GGCGCUUCAUUGAUAUAGGAUAGCUUUCAAUGGCUCUCUUGUAGUUUUGUAAGUACUUUCAUUUGACU--GUUUUCCUU--AAGAUACUAAUACUCCA--A---CAAU---CAU ((..(((((....(((((((.((((......)))).)))))))....)).)))..))......(((.(--(((......--................)--)---)).)---)). ( -16.55) >consensus GGCGCUUCAUUAAUAUAGGAUAGCUUUCAAAGGCUCUCUUGUAGUUUUGUAAGUACUUUCAUUUGUCU__GUUUUCUUU__AAGAUACUA______UA__A___CAAA___CAU ((..(((((....(((((((.((((......)))).)))))))....)).)))..))......................................................... (-14.53 = -14.87 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:37 2006