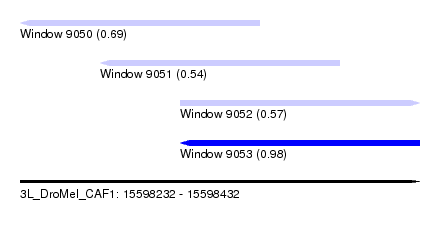
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,598,232 – 15,598,432 |
| Length | 200 |
| Max. P | 0.983226 |

| Location | 15,598,232 – 15,598,352 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -33.58 |
| Consensus MFE | -26.96 |
| Energy contribution | -27.44 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15598232 120 - 23771897 CGAGAAUUAGCUGGGCAAAUAUGCUAAGAUUUUAUAGCCUCGCAAGUGGCGGAGAUUAAACGAAACGGAGCAGAGCUCCGGAGUAAUUACAAAUGUUUUCCCUCAUUUCCACUUGGCAAA (.((......)).).......(((((((........(((.(....).)))((((((.........((((((...))))))(((..(..((....))..)..))))))))).))))))).. ( -37.10) >DroSec_CAF1 21492 120 - 1 CGAGAAUUAGCUGGGCAAAUAUGCGAAGAUUUUACAGCCUCGCAAGUGGCAGAGAUUAAACGAAACGGAGCAGAGCUCCGCAGUAAUGACAAAUGUUUUCCCUCAUUUCCACUUGGCAAA ((((.....((((...((((........))))..))))))))(((((((..(((...(((((...((((((...))))))((....)).....)))))...)))....)))))))..... ( -31.20) >DroSim_CAF1 21226 120 - 1 CGAGAAUUAGCUGGGCAAAUAUGCGAAGAUUUUACAGCCUCGCAAGUGGCAGAGAUUAAACGAAACGGAGCAGAGCUCCGGAGUAAUGACAAAUGUUUUCCCUCAUUUCCACUUGGCAAA ((((.....((((...((((........))))..))))))))(((((((.(((............((((((...))))))(((..(.(((....))).)..))).))))))))))..... ( -31.10) >DroEre_CAF1 21469 120 - 1 CGAGAAUUAGUUGGGCAAAUAUGCGGAGCUUUUACAGCCUCCAAAGUGGCGGAGAUUAAACGAAACGAGGCAGAGUUCCAGAGUAAUGGCAAAUGUUUUCCCUCAUUUCCACUUGGCAAA ((((.((((.((..(((....)))((((((((....(((((....((..((.........))..))))))))))))))).)).))))((.(((((........))))))).))))..... ( -30.10) >DroYak_CAF1 22887 119 - 1 UGAGAAUUAGCAGGGCAAAUAUGCGGAGGUUUUACAGCCUCGAAAGGGGCGGAGAUUAAACGAAACGGAGCAGAGCUCCGGAGUAAUGACAAAUGUU-UCCCUCAUUUCCACUUGGCAAA .....................(((.(((........(((((....)))))((((((.........((((((...))))))(((....(((....)))-...))))))))).))).))).. ( -38.40) >consensus CGAGAAUUAGCUGGGCAAAUAUGCGAAGAUUUUACAGCCUCGCAAGUGGCGGAGAUUAAACGAAACGGAGCAGAGCUCCGGAGUAAUGACAAAUGUUUUCCCUCAUUUCCACUUGGCAAA .....................(((.(((........(((.(....).)))((((((.........((((((...))))))(((..(.(((....))).)..))))))))).))).))).. (-26.96 = -27.44 + 0.48)

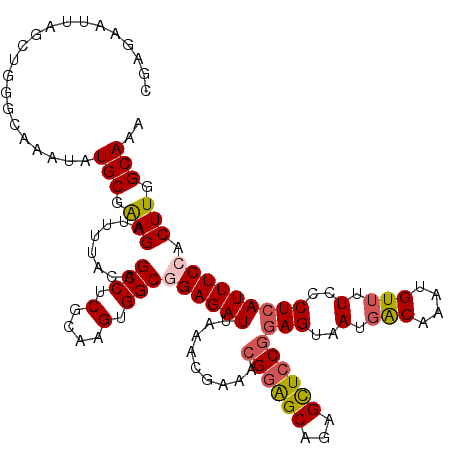
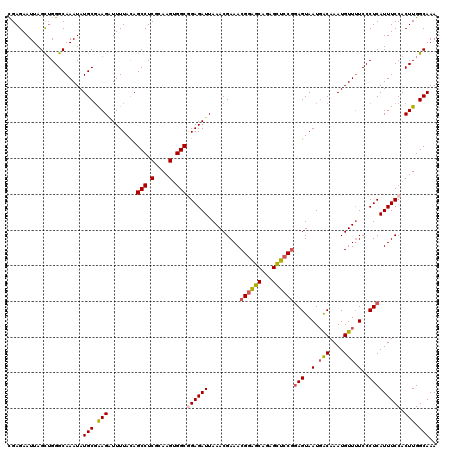
| Location | 15,598,272 – 15,598,392 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -37.54 |
| Consensus MFE | -28.94 |
| Energy contribution | -28.66 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.541084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15598272 120 - 23771897 GAGCUGCCUUGGAUUUGCCCUUAUGCACUUCACGUGUAAUCGAGAAUUAGCUGGGCAAAUAUGCUAAGAUUUUAUAGCCUCGCAAGUGGCGGAGAUUAAACGAAACGGAGCAGAGCUCCG .......(((((((((((((....((..(((.((......)).)))...)).))))))))...)))))(((((...(((.(....).))).))))).........((((((...)))))) ( -36.80) >DroSec_CAF1 21532 120 - 1 GAGCUGCCUUGGAUUUGCCCUUAUGCACUUCACGUGUAAUCGAGAAUUAGCUGGGCAAAUAUGCGAAGAUUUUACAGCCUCGCAAGUGGCAGAGAUUAAACGAAACGGAGCAGAGCUCCG ...(((((....((((((((....((..(((.((......)).)))...)).)))))))).(((((.(.........).)))))...))))).............((((((...)))))) ( -37.40) >DroSim_CAF1 21266 120 - 1 GAGCUGCCUUGGAUUUGCCCUUAUGCACUUCACGUGUAAUCGAGAAUUAGCUGGGCAAAUAUGCGAAGAUUUUACAGCCUCGCAAGUGGCAGAGAUUAAACGAAACGGAGCAGAGCUCCG ...(((((....((((((((....((..(((.((......)).)))...)).)))))))).(((((.(.........).)))))...))))).............((((((...)))))) ( -37.40) >DroEre_CAF1 21509 120 - 1 GGGCUGCCUUGGAUUUAGCCAUAUGCACUUCACGUGUAAUCGAGAAUUAGUUGGGCAAAUAUGCGGAGCUUUUACAGCCUCCAAAGUGGCGGAGAUUAAACGAAACGAGGCAGAGUUCCA ((((((((((((((((.(((((.(((((.....)))))................(((....)))((((((.....)).))))...)))))..)))))........))))))))...))). ( -34.20) >DroYak_CAF1 22926 120 - 1 GGGCUGCCUUGGAUGAGUCCAUAUGCACUUCACGUGUAAUUGAGAAUUAGCAGGGCAAAUAUGCGGAGGUUUUACAGCCUCGAAAGGGGCGGAGAUUAAACGAAACGGAGCAGAGCUCCG ....(((((((..(((.((((..(((((.....)))))..)).)).))).)))))))......((..((((((...(((((....))))).))))))...))...((((((...)))))) ( -41.90) >consensus GAGCUGCCUUGGAUUUGCCCUUAUGCACUUCACGUGUAAUCGAGAAUUAGCUGGGCAAAUAUGCGAAGAUUUUACAGCCUCGCAAGUGGCGGAGAUUAAACGAAACGGAGCAGAGCUCCG (((((..(((.(((((((((....((..(((.((......)).)))...)).)))))))).......((((((...(((.(....).))).))))))........).)))...))))).. (-28.94 = -28.66 + -0.28)

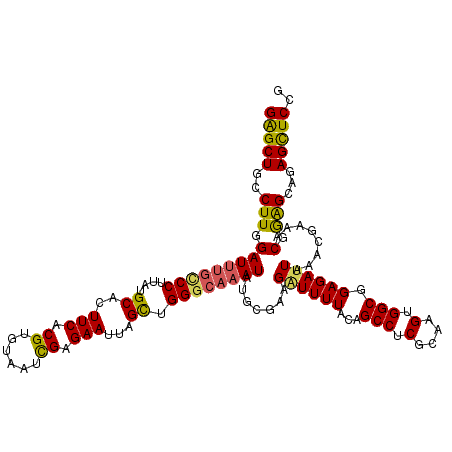
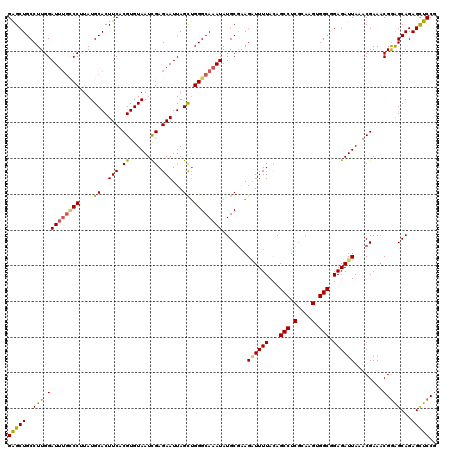
| Location | 15,598,312 – 15,598,432 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.50 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -23.52 |
| Energy contribution | -23.56 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15598312 120 + 23771897 AGGCUAUAAAAUCUUAGCAUAUUUGCCCAGCUAAUUCUCGAUUACACGUGAAGUGCAUAAGGGCAAAUCCAAGGCAGCUCACGUGUAAAACGUCGAUUUACUGAGUGAACUACUUAAUGA ..((((........))))...((..(.(((..((((..((.((((((((((..(((....(((....)))...)))..))))))))))..))..))))..))).)..))........... ( -32.50) >DroSec_CAF1 21572 120 + 1 AGGCUGUAAAAUCUUCGCAUAUUUGCCCAGCUAAUUCUCGAUUACACGUGAAGUGCAUAAGGGCAAAUCCAAGGCAGCUCACGUGUAAAAUGUCGAUUUGCUGAGUGAAUUACUUAAUGA ....................(((..(.((((......((((((((((((((..(((....(((....)))...)))..)))))))))....)))))...)))).)..))).......... ( -31.30) >DroSim_CAF1 21306 120 + 1 AGGCUGUAAAAUCUUCGCAUAUUUGCCCAGCUAAUUCUCGAUUACACGUGAAGUGCAUAAGGGCAAAUCCAAGGCAGCUCACGUGUAAAAUGUCGAUUUGCUGAGUGAAUUACUUAAUGA ....................(((..(.((((......((((((((((((((..(((....(((....)))...)))..)))))))))....)))))...)))).)..))).......... ( -31.30) >DroEre_CAF1 21549 110 + 1 AGGCUGUAAAAGCUCCGCAUAUUUGCCCAACUAAUUCUCGAUUACACGUGAAGUGCAUAUGGCUAAAUCCAAGGCAGCCCACGUGUAAGAUGUCGAUUUCCAGAGGGAAU---------- .((((.....))))..(((....)))((..((.....(((((((((((((...(((...(((......)))..)))...))))))))....))))).....))..))...---------- ( -28.20) >DroYak_CAF1 22966 120 + 1 AGGCUGUAAAACCUCCGCAUAUUUGCCCUGCUAAUUCUCAAUUACACGUGAAGUGCAUAUGGACUCAUCCAAGGCAGCCCACGUGUAAAAUGUCGAUUUUCAGAGUGAAUUACUUUAUGA ..((.(........).))..(((..(.(((..((((..((.(((((((((...(((...((((....))))..)))...)))))))))..))..))))..))).)..))).......... ( -28.10) >consensus AGGCUGUAAAAUCUUCGCAUAUUUGCCCAGCUAAUUCUCGAUUACACGUGAAGUGCAUAAGGGCAAAUCCAAGGCAGCUCACGUGUAAAAUGUCGAUUUGCUGAGUGAAUUACUUAAUGA ..((.(........).))..(((..(.(((..((((..((.((((((((((..(((....((......))...)))..))))))))))..))..))))..))).)..))).......... (-23.52 = -23.56 + 0.04)

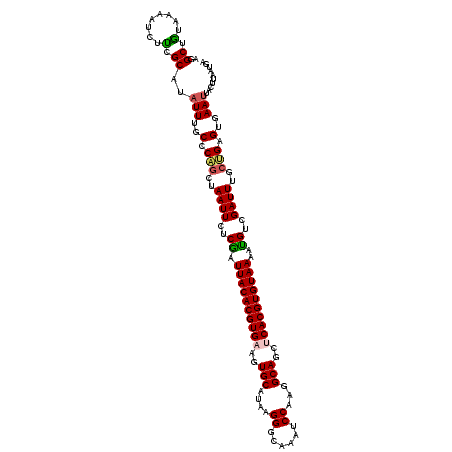
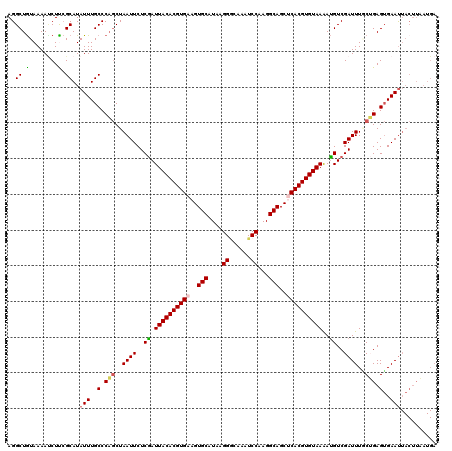
| Location | 15,598,312 – 15,598,432 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.50 |
| Mean single sequence MFE | -33.14 |
| Consensus MFE | -27.54 |
| Energy contribution | -27.10 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15598312 120 - 23771897 UCAUUAAGUAGUUCACUCAGUAAAUCGACGUUUUACACGUGAGCUGCCUUGGAUUUGCCCUUAUGCACUUCACGUGUAAUCGAGAAUUAGCUGGGCAAAUAUGCUAAGAUUUUAUAGCCU .......(((((((((...(((((.......)))))..)))))))))(((((((((((((....((..(((.((......)).)))...)).))))))))...)))))............ ( -33.30) >DroSec_CAF1 21572 120 - 1 UCAUUAAGUAAUUCACUCAGCAAAUCGACAUUUUACACGUGAGCUGCCUUGGAUUUGCCCUUAUGCACUUCACGUGUAAUCGAGAAUUAGCUGGGCAAAUAUGCGAAGAUUUUACAGCCU .......((((.((..(((((...((((....(((((((((((.(((...((......))....))).)))))))))))))))......)))))(((....)))...))..))))..... ( -33.40) >DroSim_CAF1 21306 120 - 1 UCAUUAAGUAAUUCACUCAGCAAAUCGACAUUUUACACGUGAGCUGCCUUGGAUUUGCCCUUAUGCACUUCACGUGUAAUCGAGAAUUAGCUGGGCAAAUAUGCGAAGAUUUUACAGCCU .......((((.((..(((((...((((....(((((((((((.(((...((......))....))).)))))))))))))))......)))))(((....)))...))..))))..... ( -33.40) >DroEre_CAF1 21549 110 - 1 ----------AUUCCCUCUGGAAAUCGACAUCUUACACGUGGGCUGCCUUGGAUUUAGCCAUAUGCACUUCACGUGUAAUCGAGAAUUAGUUGGGCAAAUAUGCGGAGCUUUUACAGCCU ----------...((..((((...((((....(((((((((((.(((..(((......)))...))).)))))))))))))))...))))..))(((....)))((.(((.....))))) ( -31.90) >DroYak_CAF1 22966 120 - 1 UCAUAAAGUAAUUCACUCUGAAAAUCGACAUUUUACACGUGGGCUGCCUUGGAUGAGUCCAUAUGCACUUCACGUGUAAUUGAGAAUUAGCAGGGCAAAUAUGCGGAGGUUUUACAGCCU .......((((((..(((((..((((..((..(((((((((((.(((..((((....))))...))).))))))))))).)).).)))..)))))..))).)))..((((......)))) ( -33.70) >consensus UCAUUAAGUAAUUCACUCAGCAAAUCGACAUUUUACACGUGAGCUGCCUUGGAUUUGCCCUUAUGCACUUCACGUGUAAUCGAGAAUUAGCUGGGCAAAUAUGCGAAGAUUUUACAGCCU ..........(((..((((((...((((....(((((((((((.(((...((......))....))).)))))))))))))))......))))))..))).................... (-27.54 = -27.10 + -0.44)

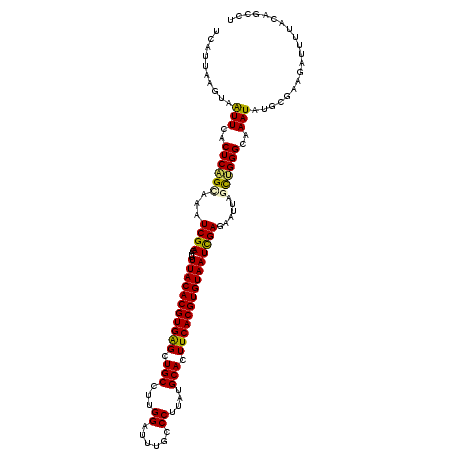
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:37 2006