
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,597,730 – 15,597,929 |
| Length | 199 |
| Max. P | 0.805418 |

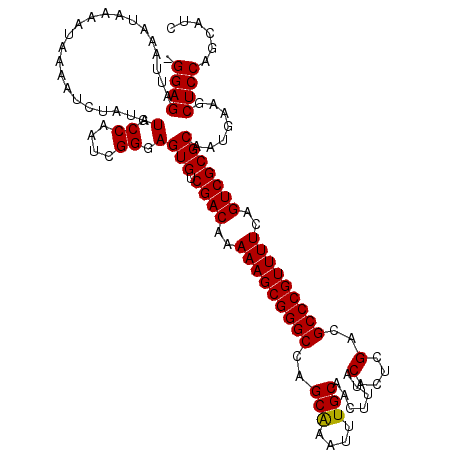
| Location | 15,597,730 – 15,597,849 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -17.58 |
| Energy contribution | -17.57 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15597730 119 - 23771897 UCGACGCCCGUUUUCAGUCGCGCAAUGAAGCUCCAGCAUCGCGGCUGAGUUUAAGAAAGAAAUAUAUACGAAAGCACAUGUGCCGACUGUACUAAAACUA-GUGACAAAUAAAUAUUACA ....(((..(((((((((((.((((((((((((.(((......)))))))))................(....)..))).)))))))))....)))))..-)))................ ( -28.00) >DroSec_CAF1 20990 119 - 1 UCGACGCCCGUUUUCAGUCGCGCAAUGAAGCUCCAGCAUCGCGGCUGAGUUAAAAAAAAAAAUAUAUACGAAAGCACAUGUGCCGACUGUACUAAAACUAAGUGAUAAA-AAAUAUUACA .........(((((((((((.((((((.(((((.(((......)))))))).................(....)..))).)))))))))....)))))...((((((..-...)))))). ( -27.70) >DroEre_CAF1 20993 95 - 1 UCGACGCCCGUUUUCAGUCGCGCAAUGAAGCUCCAGCAUCGCGGCUGAGUUCAAGAAAGAAAUAUA--CAA-----------------------CGACUAAGUGACGAAUAACUACUACA (((.(((.((((((((((((((...((......))....))))))))))(((......))).....--.))-----------------------)).....))).)))............ ( -20.50) >DroYak_CAF1 22384 118 - 1 UCGCCGCCCGUUUUCAGUCGCGCAAUGAAGCUCCAGCAUCGCGGCUGAGUUCAAGAAAGAAAUAUA--CGAAAGCACAUGUGCCAGCCGUACUAAAACUAAGUGGCAAAUAAAUACUACA ..(((((..(..((((((((((...((......))....))))))))))..)............((--((...(((....)))....))))..........))))).............. ( -31.20) >consensus UCGACGCCCGUUUUCAGUCGCGCAAUGAAGCUCCAGCAUCGCGGCUGAGUUCAAGAAAGAAAUAUA__CGAAAGCACAUGUGCCGACUGUACUAAAACUAAGUGACAAAUAAAUACUACA ....(((..(((((((((((((...((......))....))))))))))........................(((....)))............)))...)))................ (-17.58 = -17.57 + -0.00)

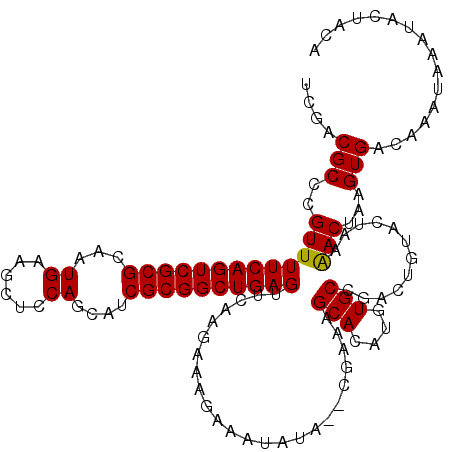

| Location | 15,597,809 – 15,597,929 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.93 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -30.85 |
| Energy contribution | -30.60 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15597809 120 - 23771897 UGGAGAUUAAAUAAAAUAAAAUCUAUAUGCCAAUCGGGAGUGUCGACAAAAAGCGGGCCAGCGAAUUUGCAACUUUACCCUCGACGCCCGUUUUCAGUCGCGCAAUGAAGCUCCAGCAUC (((((......................(.((....)).)(((.((((..(((((((((...(((................)))..)))))))))..))))))).......)))))..... ( -32.89) >DroSec_CAF1 21069 119 - 1 -GGAGAUUAAAUAAAAUAAAAUCUUUAUGCCAAUUGGGAGUGUCGACAAAAAGCGGGCCAGCGAAUUUGCAACUUUACACUCGACGCCCGUUUUCAGUCGCGCAAUGAAGCUCCAGCAUC -(((((((......))).....((((((.((....))..(((.((((..(((((((((...(((...((........)).)))..)))))))))..))))))).))))))))))...... ( -32.90) >DroEre_CAF1 21048 119 - 1 -GGAGAUUAAAUAAAAUAAAAUCUAUAUGCCAAUCGGGAGUGUCGACAAAAAGCGGGCCAGCAAAUUUGCAACUUUACACUCGACGCCCGUUUUCAGUCGCGCAAUGAAGCUCCAGCAUC -((((......................(.((....)).)(((.((((..(((((((((..(((....))).......(....)..)))))))))..))))))).......))))...... ( -31.20) >DroYak_CAF1 22462 119 - 1 -GGAGAUUAAAUAAAAUAAAAUCGAUAUGCCAAUCGGGAGUGUCGACAAAAAGCGGGCCAGCAAAUUUGCAACUUUACACUCGCCGCCCGUUUUCAGUCGCGCAAUGAAGCUCCAGCAUC -((((................(((((......)))))..(((.((((..(((((((((..((....................)).)))))))))..))))))).......))))...... ( -32.85) >consensus _GGAGAUUAAAUAAAAUAAAAUCUAUAUGCCAAUCGGGAGUGUCGACAAAAAGCGGGCCAGCAAAUUUGCAACUUUACACUCGACGCCCGUUUUCAGUCGCGCAAUGAAGCUCCAGCAUC .((((......................(.((....)).)(((.((((..(((((((((..(((....))).......(....)..)))))))))..))))))).......))))...... (-30.85 = -30.60 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:34 2006