
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
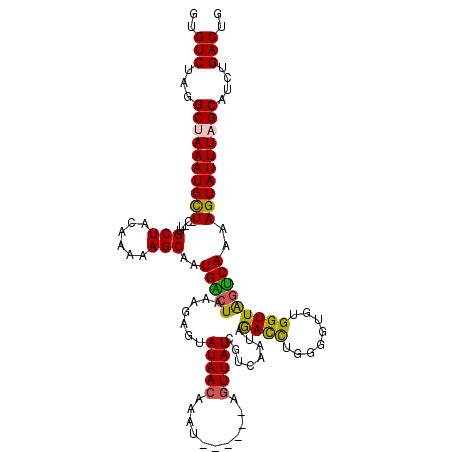
| Location | 15,594,956 – 15,595,070 |
| Length | 114 |
| Max. P | 0.630809 |

| Location | 15,594,956 – 15,595,070 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.95 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -20.50 |
| Energy contribution | -19.78 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15594956 114 + 23771897 GUGUCUAGGCUAAAUGCUUCAUUGCUACAAAAAGCACUGACUAAAGAGUAUAACAAAU------AGUUAUCGUCAAUAGACCUGGGGUGUGGUUAGUCAAAAGUAUUUAGCAACUGACUG ..(((...(((((((((((...((((......)))).(((((((.((..(((((....------.)))))..))..((.(((...))).)).))))))).)))))))))))....))).. ( -31.70) >DroSec_CAF1 18198 114 + 1 GUGUCUAGGCUAAAUGCUACAUUGCUACAAAAAGCAAUGACUAAAGUGUAUAACAAAU------AGUUAUCGUCAAUAGAUUUGGGGUGUGGUUAGUCAAAAGUAUUUAGCAUCUGACUG ..(((...((((((((((..((((((......))))))((((((..(..((..(((((------.(((......)))..)))))..))..).))))))...))))))))))....))).. ( -31.30) >DroSim_CAF1 18240 120 + 1 GUGUCUAGGCUAAAUGCUACUUUGCUACAAAAAGCAAUGACUAAAGAGUAUAACAAAUAACAAAAGUUAUCUUCAAUAGAUUUGGGGUGUGGUUAGUCAAAAGUAUUUAGCAUCUGACUG ..(((...((((((((((...(((((......)))))(((((((...((((..(((((.....(((....)))......)))))..))))..)))))))..))))))))))....))).. ( -32.60) >DroEre_CAF1 18394 110 + 1 GUGUCUAGGCAAAAUGUU---CAGCUACGAAAAGCAAUGGCUAAAAAGUAUAACAAA-------UUUUAUCGUCAAUAAACCUAGGGUGUGGUUGGUCAAAAGUAUUUAGCAUCUGACUG ..(((...((.(((((.(---(((((((.........((((...(((((.......)-------))))...))))....(((...))))))))))).......))))).))....))).. ( -16.90) >DroYak_CAF1 19610 110 + 1 GUGUCUAAGCGAAAUGUU---CAGCUACAAAAAGCAAUGGCUAAAAAUUAUAACAAA-------AGUUAUCGUCAAUAAACCAGGGGUGUGGUUAUCCAAAAGUAUUUAGCAUCUGACUG ..(((...((....(((.---.....)))....))....((((((.((((((((...-------.)))))..............(((((....)))))...))).))))))....))).. ( -17.60) >consensus GUGUCUAGGCUAAAUGCU_C_UUGCUACAAAAAGCAAUGACUAAAGAGUAUAACAAAU______AGUUAUCGUCAAUAGACCUGGGGUGUGGUUAGUCAAAAGUAUUUAGCAUCUGACUG ..(((...((((((((((.....(((......)))..(((((.......(((((...........)))))........((((........)))))))))..))))))))))....))).. (-20.50 = -19.78 + -0.72)

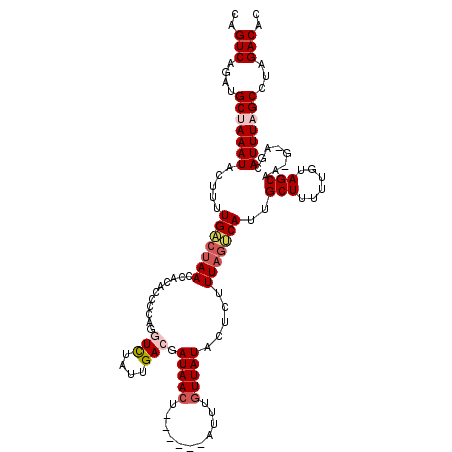
| Location | 15,594,956 – 15,595,070 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.95 |
| Mean single sequence MFE | -22.76 |
| Consensus MFE | -14.80 |
| Energy contribution | -15.92 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15594956 114 - 23771897 CAGUCAGUUGCUAAAUACUUUUGACUAACCACACCCCAGGUCUAUUGACGAUAACU------AUUUGUUAUACUCUUUAGUCAGUGCUUUUUGUAGCAAUGAAGCAUUUAGCCUAGACAC ..(((((..(((((((.(((((((((((...........(((....))).(((((.------....))))).....))))))).((((......))))..)))).))))))))).))).. ( -28.00) >DroSec_CAF1 18198 114 - 1 CAGUCAGAUGCUAAAUACUUUUGACUAACCACACCCCAAAUCUAUUGACGAUAACU------AUUUGUUAUACACUUUAGUCAUUGCUUUUUGUAGCAAUGUAGCAUUUAGCCUAGACAC ..(((((..(((((((.((...((((((........(((.....)))...(((((.------....))))).....))))))((((((......))))))..)).))))))))).))).. ( -23.30) >DroSim_CAF1 18240 120 - 1 CAGUCAGAUGCUAAAUACUUUUGACUAACCACACCCCAAAUCUAUUGAAGAUAACUUUUGUUAUUUGUUAUACUCUUUAGUCAUUGCUUUUUGUAGCAAAGUAGCAUUUAGCCUAGACAC ..(((((..(((((((.((..(((((((............((....))(((((((....)))))))..........)))))))(((((......)))))...)).))))))))).))).. ( -25.10) >DroEre_CAF1 18394 110 - 1 CAGUCAGAUGCUAAAUACUUUUGACCAACCACACCCUAGGUUUAUUGACGAUAAAA-------UUUGUUAUACUUUUUAGCCAUUGCUUUUCGUAGCUG---AACAUUUUGCCUAGACAC ..((((((...........))))))..........((((((....((((((.....-------.)))))).....((((((...(((.....)))))))---))......)))))).... ( -19.70) >DroYak_CAF1 19610 110 - 1 CAGUCAGAUGCUAAAUACUUUUGGAUAACCACACCCCUGGUUUAUUGACGAUAACU-------UUUGUUAUAAUUUUUAGCCAUUGCUUUUUGUAGCUG---AACAUUUCGCUUAGACAC ..(((.(((((((((...........(((((......)))))...((((((.....-------.)))))).....))))).))))(((......)))..---.............))).. ( -17.70) >consensus CAGUCAGAUGCUAAAUACUUUUGACUAACCACACCCCAGGUCUAUUGACGAUAACU______AUUUGUUAUACUCUUUAGUCAUUGCUUUUUGUAGCAA_G_AGCAUUUAGCCUAGACAC ..(((....(((((((.....(((((((...........(((....))).(((((...........))))).....)))))))..(((......)))........)))))))...))).. (-14.80 = -15.92 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:25 2006