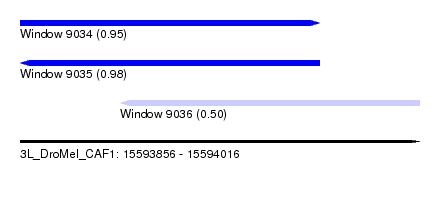
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,593,856 – 15,594,016 |
| Length | 160 |
| Max. P | 0.977232 |

| Location | 15,593,856 – 15,593,976 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -20.96 |
| Consensus MFE | -17.37 |
| Energy contribution | -18.05 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15593856 120 + 23771897 UGCAUCGCCAUUGCCAUUGCAAUUUGCACUUUAAUAAACCUGAUAGAGCACCGCAAAUGUUAAUUACAUUUGCAAAUGCAAUCAAUAUCGAACUGCCUAAUUUAAUUAACCAAUCGCUAA .((((((..((((..((((((...(((.(((((.......))).)).)))..((((((((.....))))))))...))))))))))..)))..)))........................ ( -23.10) >DroSec_CAF1 17175 120 + 1 UGCAUCGCCAUUGCCAUUGCAAUUUGCACUUUAAUAAACCUGAUAAAGCACCGCAAAUGUUAAUUACAUUUGCAAAUGCAAUCAAUAUCGAACUGCCUAAUUUAAUUAACCAAUCGCUAA .((((((..((((..((((((.......(((((.((....)).)))))....((((((((.....))))))))...))))))))))..)))..)))........................ ( -22.00) >DroSim_CAF1 17180 120 + 1 UGCAUCACCAUUGCCAUUGCAAUUUGCACUUUAAUAAACCCGAUAAAGCACCGCAAAUGUUAAUUACAUUUGCAAAUGCAAUCAAUAUCGAACUGCCUAAUUUAAUUAACCAAUCGCUAA ((((.....(((((....))))).))))..................(((...((((((((.....))))))))....(((.((......))..)))...................))).. ( -20.00) >DroEre_CAF1 17315 120 + 1 UGCAUCGCCAUUGCCAUUGCAAUUUGCACUUUAAUAAACCUGAUAAAGCACCCCAAAUGUUAAUUACUUUUGCAAAUGCAAUCAGUAUCGAACUGCCUAAUUUAAUUAACCAAUCGCUAA .((((((..((((..((((((.(((((((((((.((....)).)))))..........((.....))...))))))))))))))))..)))..)))........................ ( -17.00) >DroYak_CAF1 18531 120 + 1 UGCAUCGCCGUUGCCAUUGCAAUUUGCACUGUAAUAAACCUGAUGAAGCACCGCAAAUGUUAAUUACUUUUGCAAAUGCAAUCAAUAUCGAACAGCCUAAUUGAAUUAACCAAUCGCUAA ....(((..((((..((((((.((((((..(((((.(((.((.((......))))...))).)))))...))))))))))))))))..)))..(((...((((.......)))).))).. ( -22.70) >consensus UGCAUCGCCAUUGCCAUUGCAAUUUGCACUUUAAUAAACCUGAUAAAGCACCGCAAAUGUUAAUUACAUUUGCAAAUGCAAUCAAUAUCGAACUGCCUAAUUUAAUUAACCAAUCGCUAA .((((((..((((..((((((...(((....................)))..((((((((.....))))))))...))))))))))..)))..)))........................ (-17.37 = -18.05 + 0.68)

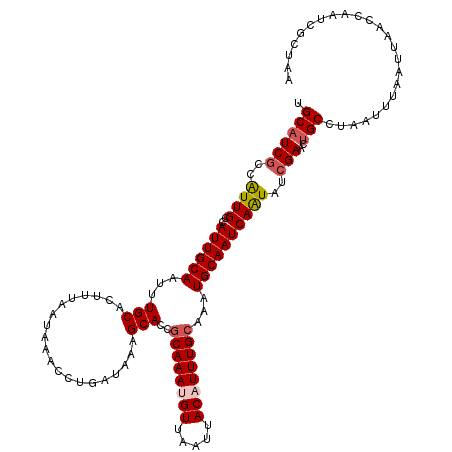
| Location | 15,593,856 – 15,593,976 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -26.48 |
| Energy contribution | -27.20 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15593856 120 - 23771897 UUAGCGAUUGGUUAAUUAAAUUAGGCAGUUCGAUAUUGAUUGCAUUUGCAAAUGUAAUUAACAUUUGCGGUGCUCUAUCAGGUUUAUUAAAGUGCAAAUUGCAAUGGCAAUGGCGAUGCA ...((.((((..............(((.((..((((((((.(((((.((((((((.....)))))))))))))...)))))...)))..)).)))..(((((....)))))..)))))). ( -32.10) >DroSec_CAF1 17175 120 - 1 UUAGCGAUUGGUUAAUUAAAUUAGGCAGUUCGAUAUUGAUUGCAUUUGCAAAUGUAAUUAACAUUUGCGGUGCUUUAUCAGGUUUAUUAAAGUGCAAAUUGCAAUGGCAAUGGCGAUGCA ...(((((((.(((((...))))).))))(((.(((((((((((..(((((((((.....)))))))))(..(((((..........)))))..)....))))))..))))).)))))). ( -32.50) >DroSim_CAF1 17180 120 - 1 UUAGCGAUUGGUUAAUUAAAUUAGGCAGUUCGAUAUUGAUUGCAUUUGCAAAUGUAAUUAACAUUUGCGGUGCUUUAUCGGGUUUAUUAAAGUGCAAAUUGCAAUGGCAAUGGUGAUGCA ...(((((((.(((((...))))).))))(((.(((((((((((..(((((((((.....)))))))))(..(((((..........)))))..)....))))))..))))).)))))). ( -30.90) >DroEre_CAF1 17315 120 - 1 UUAGCGAUUGGUUAAUUAAAUUAGGCAGUUCGAUACUGAUUGCAUUUGCAAAAGUAAUUAACAUUUGGGGUGCUUUAUCAGGUUUAUUAAAGUGCAAAUUGCAAUGGCAAUGGCGAUGCA ...((.((((..............(((.((..((((((((.((((((.((((.((.....)).))))))))))...)))))...)))..)).)))..(((((....)))))..)))))). ( -27.00) >DroYak_CAF1 18531 120 - 1 UUAGCGAUUGGUUAAUUCAAUUAGGCUGUUCGAUAUUGAUUGCAUUUGCAAAAGUAAUUAACAUUUGCGGUGCUUCAUCAGGUUUAUUACAGUGCAAAUUGCAAUGGCAACGGCGAUGCA ...((((((((.....))))))..(((((......(((((.(((((.(((((.((.....)).))))))))))...))))).......)))))))..(((((...(....).)))))... ( -29.12) >consensus UUAGCGAUUGGUUAAUUAAAUUAGGCAGUUCGAUAUUGAUUGCAUUUGCAAAUGUAAUUAACAUUUGCGGUGCUUUAUCAGGUUUAUUAAAGUGCAAAUUGCAAUGGCAAUGGCGAUGCA ...((.((((..............(((.((..((((((((.(((((.((((((((.....)))))))))))))...)))))...)))..)).)))..(((((....)))))..)))))). (-26.48 = -27.20 + 0.72)

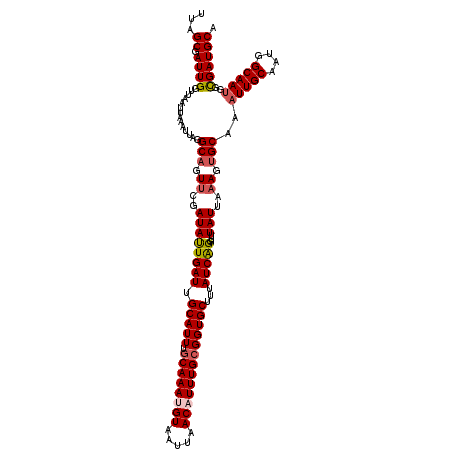

| Location | 15,593,896 – 15,594,016 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -21.06 |
| Energy contribution | -21.46 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15593896 120 - 23771897 CCUACAAUUUUAAUCAACUUGGCCACGCGGCAAAUAAAACUUAGCGAUUGGUUAAUUAAAUUAGGCAGUUCGAUAUUGAUUGCAUUUGCAAAUGUAAUUAACAUUUGCGGUGCUCUAUCA ...........((((((....(((....))).............((((((.(((((...))))).))).)))...))))))(((((.((((((((.....)))))))))))))....... ( -26.70) >DroSec_CAF1 17215 120 - 1 CCUACAAUUUUAAUCAACUUGGCCACGCGGCAAAUAAAAUUUAGCGAUUGGUUAAUUAAAUUAGGCAGUUCGAUAUUGAUUGCAUUUGCAAAUGUAAUUAACAUUUGCGGUGCUUUAUCA ...........((((((....(((....))).............((((((.(((((...))))).))).)))...))))))(((((.((((((((.....)))))))))))))....... ( -26.70) >DroSim_CAF1 17220 120 - 1 CCUACAAUUUUAAUCAACUUGGCCACGCGGCAAAUAAAACUUAGCGAUUGGUUAAUUAAAUUAGGCAGUUCGAUAUUGAUUGCAUUUGCAAAUGUAAUUAACAUUUGCGGUGCUUUAUCG ...........((((((....(((....))).............((((((.(((((...))))).))).)))...))))))(((((.((((((((.....)))))))))))))....... ( -26.70) >DroEre_CAF1 17355 120 - 1 CCCACAAUUUUAAUCAACUUGACCAGGCCGCAAAUAAACCUUAGCGAUUGGUUAAUUAAAUUAGGCAGUUCGAUACUGAUUGCAUUUGCAAAAGUAAUUAACAUUUGGGGUGCUUUAUCA ((((....((((((....((((((((..(((............))).))))))))....))))))..(((.(.((((..((((....)))).)))).).)))...))))........... ( -24.40) >DroYak_CAF1 18571 120 - 1 CCUACCAUUUUAAUCAACUUGACCACGCCGCAAAUAAAACUUAGCGAUUGGUUAAUUCAAUUAGGCUGUUCGAUAUUGAUUGCAUUUGCAAAAGUAAUUAACAUUUGCGGUGCUUCAUCA ...................(((...((((((((((......((((((((((.....))))))..)))).....((((..((((....)))).))))......))))))))))..)))... ( -25.60) >consensus CCUACAAUUUUAAUCAACUUGGCCACGCGGCAAAUAAAACUUAGCGAUUGGUUAAUUAAAUUAGGCAGUUCGAUAUUGAUUGCAUUUGCAAAUGUAAUUAACAUUUGCGGUGCUUUAUCA ........((((((....((((((((((...............)))..)))))))....))))))...........((((.(((((.((((((((.....)))))))))))))...)))) (-21.06 = -21.46 + 0.40)

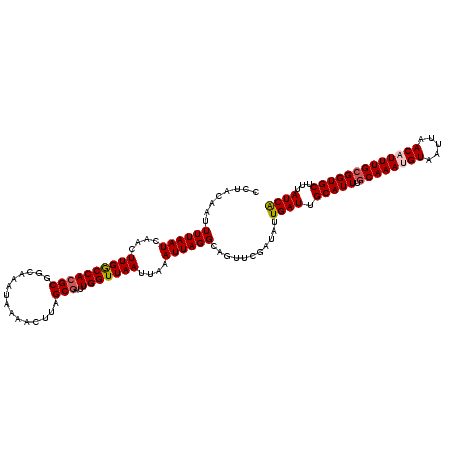
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:20 2006