
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,590,372 – 15,590,647 |
| Length | 275 |
| Max. P | 0.997413 |

| Location | 15,590,372 – 15,590,488 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.94 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -25.16 |
| Energy contribution | -25.17 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15590372 116 + 23771897 AUAAGGGAAAGCAAAAAGACAGUCAACGUGACACACAACACAUAGUCACACAAACACAC----ACAUGUGCACGGGAGUUGUGCAUGUUGGUGUGUGCGAAAAUAUGCAAAUUAAGUGUG .....................(((.....)))..............(((((..((((((----(((((..(((....).))..)))))..))))))(((......))).......))))) ( -28.60) >DroSec_CAF1 13624 118 + 1 AUAAGGGAAAGCAAGAAGACAGUCAACGUGACACACAACACAUAGGCACACAAACACAC--ACACGUGUGCACGGGAGUUGUGCAUGCUGGUGUGUGCGAAAAUAUGCAAAUUAAGUGUG .....................(((.....)))..............(((((..((((((--.((.(((((((((.....)))))))))))))))))(((......))).......))))) ( -30.10) >DroYak_CAF1 14912 116 + 1 GUAAGGGAAAGCCAAAAGAUAGUCGACGUGACACACAACACAUAGGCACA----CACACACACGCAUGUGCACGGGAGUUGUGCAUGUGGGUGUGUGCGAAAAUAUGCAAAUUAAGUGUG ..........(((....(.....)...(((........)))...))).((----(((((((((.((((((((((.....)))))))))).))))))(((......))).......))))) ( -36.60) >consensus AUAAGGGAAAGCAAAAAGACAGUCAACGUGACACACAACACAUAGGCACACAAACACAC__ACACAUGUGCACGGGAGUUGUGCAUGUUGGUGUGUGCGAAAAUAUGCAAAUUAAGUGUG .....................(((.....))).....((((.............(((((....(((((..(((....).))..)))))..)))))((((......))))......)))). (-25.16 = -25.17 + 0.00)

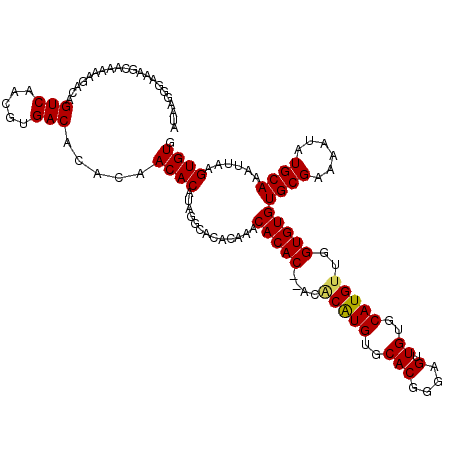
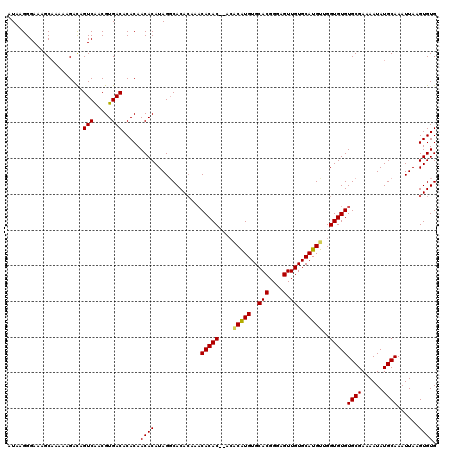
| Location | 15,590,372 – 15,590,488 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.94 |
| Mean single sequence MFE | -33.11 |
| Consensus MFE | -22.23 |
| Energy contribution | -23.57 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15590372 116 - 23771897 CACACUUAAUUUGCAUAUUUUCGCACACACCAACAUGCACAACUCCCGUGCACAUGU----GUGUGUUUGUGUGACUAUGUGUUGUGUGUCACGUUGACUGUCUUUUUGCUUUCCCUUAU .(((.(((((.(((((((...(((((((((.((((((((((.............)))----))))))).)))))....))))..))))).)).))))).))).................. ( -29.52) >DroSec_CAF1 13624 118 - 1 CACACUUAAUUUGCAUAUUUUCGCACACACCAGCAUGCACAACUCCCGUGCACACGUGU--GUGUGUUUGUGUGCCUAUGUGUUGUGUGUCACGUUGACUGUCUUCUUGCUUUCCCUUAU (((((...((..((((((...(((((((((.....(((((.......)))))...))))--)))))...))))))..)).....)))))............................... ( -33.10) >DroYak_CAF1 14912 116 - 1 CACACUUAAUUUGCAUAUUUUCGCACACACCCACAUGCACAACUCCCGUGCACAUGCGUGUGUGUG----UGUGCCUAUGUGUUGUGUGUCACGUCGACUAUCUUUUGGCUUUCCCUUAC (((((.......((((((....((((((((.((((((((..((....)).....)))))))).)))----)))))..)))))).)))))....(((((.......))))).......... ( -36.70) >consensus CACACUUAAUUUGCAUAUUUUCGCACACACCAACAUGCACAACUCCCGUGCACAUGUGU__GUGUGUUUGUGUGCCUAUGUGUUGUGUGUCACGUUGACUGUCUUUUUGCUUUCCCUUAU .(((.(((((.(((((((...(((((((((.(((..((((.......))))((((......))))))).)))))....))))..))))).)).))))).))).................. (-22.23 = -23.57 + 1.34)



| Location | 15,590,412 – 15,590,528 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -41.07 |
| Consensus MFE | -34.60 |
| Energy contribution | -35.27 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15590412 116 + 23771897 CAUAGUCACACAAACACAC----ACAUGUGCACGGGAGUUGUGCAUGUUGGUGUGUGCGAAAAUAUGCAAAUUAAGUGUGGCGACUUUUGUUCUCGCCGUUUUUUGGUGUAGAAAGCGUA ....(((((((..((((((----(((((..(((....).))..)))))..))))))(((......))).......)))))))........(((((((((.....))))).))))...... ( -40.30) >DroSec_CAF1 13664 118 + 1 CAUAGGCACACAAACACAC--ACACGUGUGCACGGGAGUUGUGCAUGCUGGUGUGUGCGAAAAUAUGCAAAUUAAGUGUGGCGACUUUUGUUUUCGCCGUUUUUUGGUAUAGAAAGCGUA ....(.(((((..((((((--.((.(((((((((.....)))))))))))))))))(((......))).......))))).).......((((((((((.....))))...))))))... ( -39.20) >DroYak_CAF1 14952 116 + 1 CAUAGGCACA----CACACACACGCAUGUGCACGGGAGUUGUGCAUGUGGGUGUGUGCGAAAAUAUGCAAAUUAAGUGUGGCGACUUUUGUUCUCGCCGUUUUUUGGUGUAGAAAGCGUA ......((((----(((((((.((((((..(((....).))..)))))).))))))(((......))).......)))))(((.(((((.....(((((.....)))))..)))))))). ( -43.70) >consensus CAUAGGCACACAAACACAC__ACACAUGUGCACGGGAGUUGUGCAUGUUGGUGUGUGCGAAAAUAUGCAAAUUAAGUGUGGCGACUUUUGUUCUCGCCGUUUUUUGGUGUAGAAAGCGUA ......(((((...(((((....(((((..(((....).))..)))))..)))))((((......))))......)))))(((.(((((.....(((((.....)))))..)))))))). (-34.60 = -35.27 + 0.67)



| Location | 15,590,412 – 15,590,528 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -32.03 |
| Consensus MFE | -22.67 |
| Energy contribution | -23.34 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15590412 116 - 23771897 UACGCUUUCUACACCAAAAAACGGCGAGAACAAAAGUCGCCACACUUAAUUUGCAUAUUUUCGCACACACCAACAUGCACAACUCCCGUGCACAUGU----GUGUGUUUGUGUGACUAUG ...........((((((.....(((((.........))))).....................((((((((.....(((((.......)))))...))----))))))))).)))...... ( -28.30) >DroSec_CAF1 13664 118 - 1 UACGCUUUCUAUACCAAAAAACGGCGAAAACAAAAGUCGCCACACUUAAUUUGCAUAUUUUCGCACACACCAGCAUGCACAACUCCCGUGCACACGUGU--GUGUGUUUGUGUGCCUAUG ......................(((((.........)))))...........((((((...(((((((((.....(((((.......)))))...))))--)))))...))))))..... ( -34.40) >DroYak_CAF1 14952 116 - 1 UACGCUUUCUACACCAAAAAACGGCGAGAACAAAAGUCGCCACACUUAAUUUGCAUAUUUUCGCACACACCCACAUGCACAACUCCCGUGCACAUGCGUGUGUGUG----UGUGCCUAUG ......................(((((.........)))))...........(((((....(((((((((.((..(((((.......)))))..)).)))))))))----)))))..... ( -33.40) >consensus UACGCUUUCUACACCAAAAAACGGCGAGAACAAAAGUCGCCACACUUAAUUUGCAUAUUUUCGCACACACCAACAUGCACAACUCCCGUGCACAUGUGU__GUGUGUUUGUGUGCCUAUG ......................(((((.........)))))...........((((((...(((((.........(((((.......))))).........)))))...))))))..... (-22.67 = -23.34 + 0.67)



| Location | 15,590,528 – 15,590,647 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -33.67 |
| Consensus MFE | -29.88 |
| Energy contribution | -31.24 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.997413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15590528 119 + 23771897 AAAAGCAAGCAAAAACUGUACGUAUACACAUUAUUUGGGCUGGCGAUUGUGGAAAGCAAAGCGCCGAAA-AAAACGAGUAUACGCCCCGUUGUACAGUCAGUCAGUCCGCAAUAUGUGUG ....((..(((.....)))..)).(((((((.((((((((((((((((((((...((...)).))....-..((((.((....))..))))..)))))).))))))))).)))))))))) ( -31.30) >DroSec_CAF1 13782 119 + 1 AAAAGCGAGCAAAAACUGUACGUAUACACAUUAUUUGGGCUGGCGAUUGUGGAAAGCAAAGCGCCGAAA-AAAACGAUAAUACGCCCCGUUGUACAGUCAGUCAGUCCGCAAUAUGUGUA ....(((.(((.....))).))).(((((((.(((((((((((((((((((.((......(((......-............)))....)).))))))).))))))))).)))))))))) ( -34.77) >DroSim_CAF1 13745 119 + 1 AAAAGCGAGCAAAAACUGUACGUAUACACAUUAUUUGGGCUGGCGAUUGUGGAAAGCAAAGCGCCGAAA-AAAACGAGUAUACGCCCCGUUGUACAGUCAGUCAGCCCGCAAUAUGUGUG ....(((.(((.....))).))).(((((((.((((((((((((((((((((...((...)).))....-..((((.((....))..))))..)))))).))))))))).)))))))))) ( -37.70) >DroEre_CAF1 13748 110 + 1 AAAAGCGAGCAAAAACUGUACGUAUACACAUUAUUUGGGCUGGCGAUUGUGGAAAGCAAAGCGCCGAAAAAAAACGAGUAUACGCCCCGUUG---------UCAGUCCGCAGUAUGUGU- ....(((.(((.....))).)))..((((((.(((((((((((((((.(.((...((...))((((........)).)).....))).))))---------)))))))).)))))))))- ( -29.60) >DroYak_CAF1 15068 119 + 1 AAAAGCGAGCAAAAACUGUACGUAUACACAUUAUUUGGGCUGGCGAUUGUGGAAAGCAAAGCGCCGAAA-AAAACGAGUAUACGCCCCGUUGUACAGUCAGUCAGUCCGCAAUAUGUGUG ....(((.(((.....))).))).(((((((.((((((((((((((((((((...((...)).))....-..((((.((....))..))))..)))))).))))))))).)))))))))) ( -35.00) >consensus AAAAGCGAGCAAAAACUGUACGUAUACACAUUAUUUGGGCUGGCGAUUGUGGAAAGCAAAGCGCCGAAA_AAAACGAGUAUACGCCCCGUUGUACAGUCAGUCAGUCCGCAAUAUGUGUG ....(((.(((.....))).))).(((((((.((((((((((((((((((((...((...)).)).......((((...........))))..)))))).))))))))).)))))))))) (-29.88 = -31.24 + 1.36)

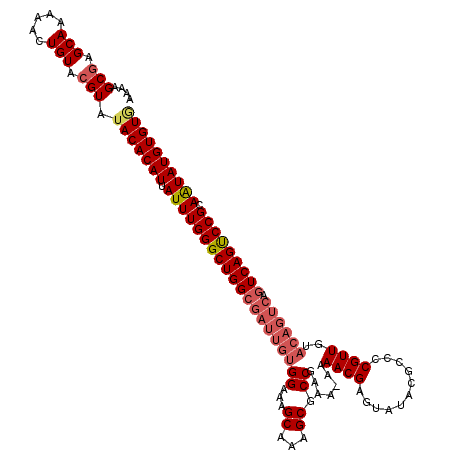
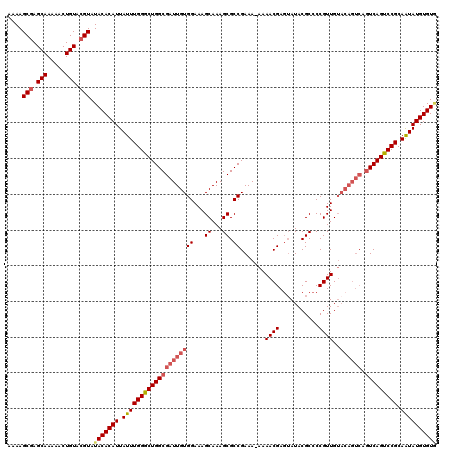
| Location | 15,590,528 – 15,590,647 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -30.94 |
| Consensus MFE | -24.65 |
| Energy contribution | -26.09 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15590528 119 - 23771897 CACACAUAUUGCGGACUGACUGACUGUACAACGGGGCGUAUACUCGUUUU-UUUCGGCGCUUUGCUUUCCACAAUCGCCAGCCCAAAUAAUGUGUAUACGUACAGUUUUUGCUUGCUUUU ..........(((((..(((((.(((.....))).((((((((.((((.(-((..((((..(((.......))).)))).....))).)))).)))))))).))))))))))........ ( -29.00) >DroSec_CAF1 13782 119 - 1 UACACAUAUUGCGGACUGACUGACUGUACAACGGGGCGUAUUAUCGUUUU-UUUCGGCGCUUUGCUUUCCACAAUCGCCAGCCCAAAUAAUGUGUAUACGUACAGUUUUUGCUCGCUUUU ..........(((..(.(((((((((((((...(((((......).....-....((((..(((.......))).)))).))))........)))))).)).)))))...)..))).... ( -30.30) >DroSim_CAF1 13745 119 - 1 CACACAUAUUGCGGGCUGACUGACUGUACAACGGGGCGUAUACUCGUUUU-UUUCGGCGCUUUGCUUUCCACAAUCGCCAGCCCAAAUAAUGUGUAUACGUACAGUUUUUGCUCGCUUUU ..........((((((.(((((.(((.....))).((((((((.((((.(-((..((((..(((.......))).)))).....))).)))).)))))))).)))))...)))))).... ( -37.00) >DroEre_CAF1 13748 110 - 1 -ACACAUACUGCGGACUGA---------CAACGGGGCGUAUACUCGUUUUUUUUCGGCGCUUUGCUUUCCACAAUCGCCAGCCCAAAUAAUGUGUAUACGUACAGUUUUUGCUCGCUUUU -.........(((..(.((---------.(((...((((((((.((((.(((...((((..(((.......))).)))).....))).)))).))))))))...))))).)..))).... ( -26.90) >DroYak_CAF1 15068 119 - 1 CACACAUAUUGCGGACUGACUGACUGUACAACGGGGCGUAUACUCGUUUU-UUUCGGCGCUUUGCUUUCCACAAUCGCCAGCCCAAAUAAUGUGUAUACGUACAGUUUUUGCUCGCUUUU ..........(((..(.(((((.(((.....))).((((((((.((((.(-((..((((..(((.......))).)))).....))).)))).)))))))).)))))...)..))).... ( -31.50) >consensus CACACAUAUUGCGGACUGACUGACUGUACAACGGGGCGUAUACUCGUUUU_UUUCGGCGCUUUGCUUUCCACAAUCGCCAGCCCAAAUAAUGUGUAUACGUACAGUUUUUGCUCGCUUUU ..........(((..(.(((((.(((.....))).((((((((.((((.......((((..(((.......))).)))).........)))).)))))))).)))))...)..))).... (-24.65 = -26.09 + 1.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:06 2006