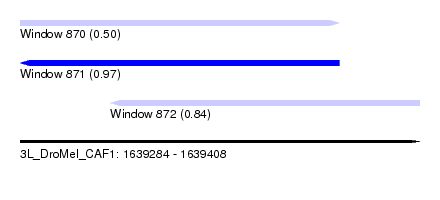
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,639,284 – 1,639,408 |
| Length | 124 |
| Max. P | 0.972234 |

| Location | 1,639,284 – 1,639,383 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 83.92 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -27.23 |
| Energy contribution | -26.78 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
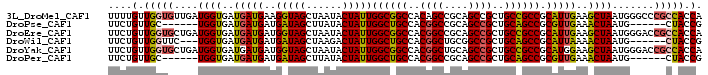
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1639284 99 + 23771897 UUUUGUUGGUGUUGAUGGUGAUGAUGAAGGUAGCUAAUACUAUUGGCGGCCACAGCCGCAGCCGCUGCCGCCGCAUUGAAGCUAAUGGGCCCGCCACCA ............((.(((((........((((((.....(....)(((((....)))))....))))))(((.(((((....)))))))).))))).)) ( -33.00) >DroPse_CAF1 1312 87 + 1 UUCUGUUGC------UGGUGAUGAUGAUGAUAGCUUAUACUAUUGGCUGCCACGGCCGCAGCCGCUGCAGCCGCGUUGAAACUAAUG------CUACCG .........------.((((........(((((......)))))((((((..((((....))))..))))))((((((....)))))------))))). ( -29.30) >DroEre_CAF1 1200 99 + 1 UUCUGUUGGUGCUGAUGGUGAUGAUGAUGGUAGCUAAUACUAUUGGCGGCCACGGCCGCAGCCGCUGCCGCCGCAUUGAAGCUAAUGGGACCGCCACCA ...((.((((.(((.((((..(((((((((((.....)))))).((((((..((((....))))..)))))).)))))..)))).))).)))).))... ( -37.90) >DroWil_CAF1 1439 90 + 1 UUCUGUUGGUUC---UGGUGAUGAUGAUGAUAGCUAAGACUAUUGGCUGCCACGGCUGCGGCCGCUGCAGCCGCAUUAAAACUAAUG------CUACCG ....(.((((..---((((..(((((.((.(((((((.....))))))).))(((((((((...))))))))))))))..))))..)------))).). ( -32.90) >DroYak_CAF1 1202 99 + 1 UUCUGUUGGUGCUGAUGGUGAUGAUGAUGGUAGCUAAUACUAUUGGCGGCCACGGCUGCAGCCGCUGCCGCCGCAUGGAAGCUAAUGGGACCGCCACCA ...((.((((.(((.((((..(.(((((((((.....)))))).((((((..((((....))))..)))))).))).)..)))).))).)))).))... ( -38.00) >DroPer_CAF1 1298 87 + 1 UUCUGUUGC------UGGUGAUGAUGAUGAUAGCUUAUACUAUUGGCUGCCACGGCCGCAGCCGCUGCAGCCGCGUUGAAACUAAUG------CUACCG .........------.((((........(((((......)))))((((((..((((....))))..))))))((((((....)))))------))))). ( -29.30) >consensus UUCUGUUGGUGC___UGGUGAUGAUGAUGAUAGCUAAUACUAUUGGCGGCCACGGCCGCAGCCGCUGCAGCCGCAUUGAAACUAAUG______CCACCA ....(.(((((....((((..(((((..(((((......)))))((((((..((((....))))..)))))).)))))..)))).......))))).). (-27.23 = -26.78 + -0.44)
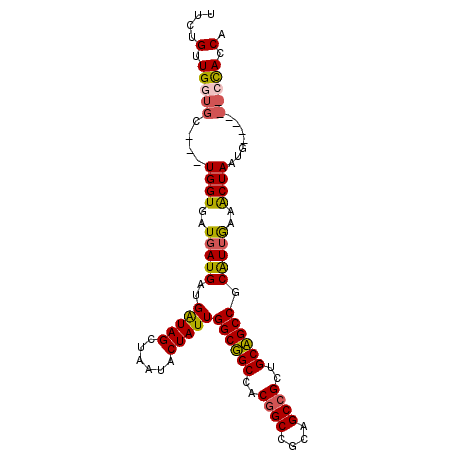
| Location | 1,639,284 – 1,639,383 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 83.92 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -30.25 |
| Energy contribution | -28.87 |
| Covariance contribution | -1.39 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1639284 99 - 23771897 UGGUGGCGGGCCCAUUAGCUUCAAUGCGGCGGCAGCGGCUGCGGCUGUGGCCGCCAAUAGUAUUAGCUACCUUCAUCAUCACCAUCAACACCAACAAAA (((((..((......(((((..(((((((((((.(((((....))))).))))))....))))))))))......))..)))))............... ( -36.20) >DroPse_CAF1 1312 87 - 1 CGGUAG------CAUUAGUUUCAACGCGGCUGCAGCGGCUGCGGCCGUGGCAGCCAAUAGUAUAAGCUAUCAUCAUCAUCACCA------GCAACAGAA .(((((------(....((......))((((((.(((((....))))).))))))..........)))))).............------......... ( -31.10) >DroEre_CAF1 1200 99 - 1 UGGUGGCGGUCCCAUUAGCUUCAAUGCGGCGGCAGCGGCUGCGGCCGUGGCCGCCAAUAGUAUUAGCUACCAUCAUCAUCACCAUCAGCACCAACAGAA (((((..(((.....(((((..(((((((((((.(((((....))))).))))))....)))))))))).....)))..)))))............... ( -38.90) >DroWil_CAF1 1439 90 - 1 CGGUAG------CAUUAGUUUUAAUGCGGCUGCAGCGGCCGCAGCCGUGGCAGCCAAUAGUCUUAGCUAUCAUCAUCAUCACCA---GAACCAACAGAA .(((.(------(((((....))))))((((((.(((((....))))).)))))).(((((....)))))..........))).---............ ( -31.60) >DroYak_CAF1 1202 99 - 1 UGGUGGCGGUCCCAUUAGCUUCCAUGCGGCGGCAGCGGCUGCAGCCGUGGCCGCCAAUAGUAUUAGCUACCAUCAUCAUCACCAUCAGCACCAACAGAA (((((..(((.....(((((...((((((((((.(((((....))))).))))))....)))).))))).....)))..)))))............... ( -38.30) >DroPer_CAF1 1298 87 - 1 CGGUAG------CAUUAGUUUCAACGCGGCUGCAGCGGCUGCGGCCGUGGCAGCCAAUAGUAUAAGCUAUCAUCAUCAUCACCA------GCAACAGAA .(((((------(....((......))((((((.(((((....))))).))))))..........)))))).............------......... ( -31.10) >consensus CGGUAG______CAUUAGCUUCAAUGCGGCGGCAGCGGCUGCGGCCGUGGCAGCCAAUAGUAUUAGCUACCAUCAUCAUCACCA___GCACCAACAGAA .(((((...........((......))((((((.(((((....))))).))))))...........)))))............................ (-30.25 = -28.87 + -1.39)

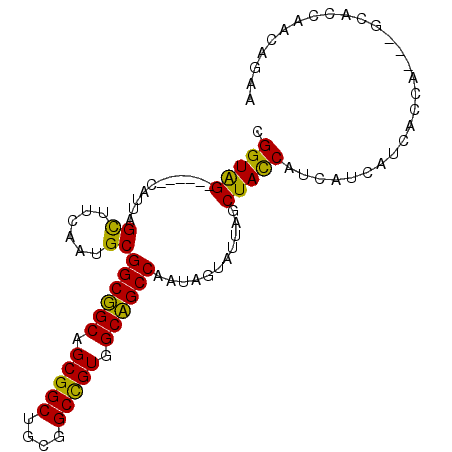
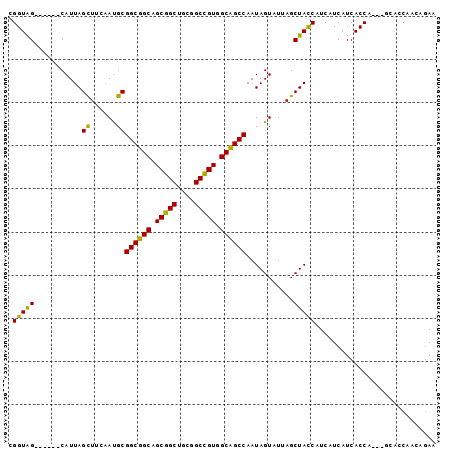
| Location | 1,639,312 – 1,639,408 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 77.19 |
| Mean single sequence MFE | -40.30 |
| Consensus MFE | -27.69 |
| Energy contribution | -27.47 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1639312 96 - 23771897 UG---GGCCAUCCGUCG---CAUGUCCGGGGUGGUGGCGGGCCCAUUAGCUUCAAUGCGGCGGCAGCGGCUGCGGCUGUGGCCGCCAAUAGUAUUAGCUACC ((---((((..((..((---(.........)))..))..)))))).(((((..(((((((((((.(((((....))))).))))))....)))))))))).. ( -46.40) >DroEre_CAF1 1228 96 - 1 UG---GGUCAUCCUUCU---CAUAUCCGGGGUGGUGGCGGUCCCAUUAGCUUCAAUGCGGCGGCAGCGGCUGCGGCCGUGGCCGCCAAUAGUAUUAGCUACC ((---((.(..((.(((---(......)))).)).....).)))).(((((..(((((((((((.(((((....))))).))))))....)))))))))).. ( -40.70) >DroWil_CAF1 1464 93 - 1 UCGGUGGUCAUCCGCAG---CAUGCGCGGGGCGGUAG------CAUUAGUUUUAAUGCGGCUGCAGCGGCCGCAGCCGUGGCAGCCAAUAGUCUUAGCUAUC ..((((((...((((..---.....))))(((....(------(((((....))))))((((((.(((((....))))).))))))....)))...)))))) ( -39.80) >DroYak_CAF1 1230 96 - 1 UG---GGUCAUCCUUCG---CAUGUCCGGGGUGGUGGCGGUCCCAUUAGCUUCCAUGCGGCGGCAGCGGCUGCAGCCGUGGCCGCCAAUAGUAUUAGCUACC ((---((((((((((((---......))))).)))))....)))).(((((...((((((((((.(((((....))))).))))))....)))).))))).. ( -41.60) >DroMoj_CAF1 1830 96 - 1 UC---GGCCAUCCGGCUCAGCAUGGGCGCGG---CACUGGCGGCAUCAGCUUCAAUGCGGCUGCAGCGGCCGCUGCGGUGGCGGCCAAUAGUAUUAGCUAUC ..---((((..(((((((.....))))((((---(....(((((....((......)).)))))....)))))..)))....)))).(((((....))))). ( -42.10) >DroPer_CAF1 1320 90 - 1 UU---AGUCAUCCCCAG---CAUGCGAGGACCGGUAG------CAUUAGUUUCAACGCGGCUGCAGCGGCUGCGGCCGUGGCAGCCAAUAGUAUAAGCUAUC ..---.(((.((.(...---...).)).))).(((((------(....((......))((((((.(((((....))))).))))))..........)))))) ( -31.20) >consensus UG___GGUCAUCCGUCG___CAUGUCCGGGGUGGUAGCGG_CCCAUUAGCUUCAAUGCGGCGGCAGCGGCUGCGGCCGUGGCCGCCAAUAGUAUUAGCUACC ................................((((((..........((......))((((((.(((((....))))).))))))..........)))))) (-27.69 = -27.47 + -0.22)

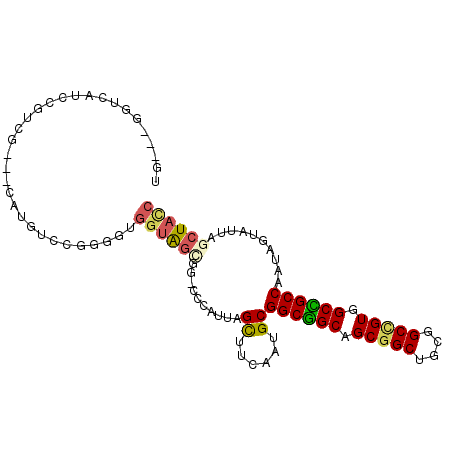
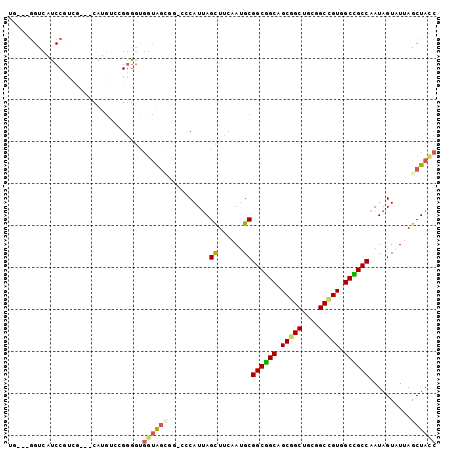
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:35 2006