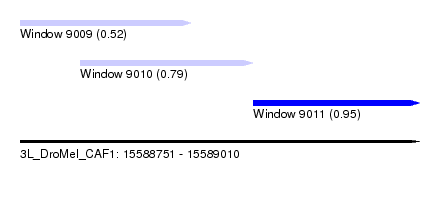
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,588,751 – 15,589,010 |
| Length | 259 |
| Max. P | 0.951486 |

| Location | 15,588,751 – 15,588,862 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.21 |
| Mean single sequence MFE | -17.15 |
| Consensus MFE | -12.22 |
| Energy contribution | -11.90 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15588751 111 + 23771897 GAAACUGAAAUUUCAUAUGAACCAAAUUGAAGUCCCA-AUCAAGGUUU---UUACGAAUAAUUUGUAAACAA----ACGCUCGACCA-AUUCAAUCUUAUCGAGCUUCAAUAUCAGCAAA ....((((...(((....)))....((((((......-......((((---((((((.....))))))..))----))((((((...-...........)))))))))))).)))).... ( -18.04) >DroSec_CAF1 12097 117 + 1 GAAACUCAAAUUUCAUUUGAACCAAAUUGAAGUCCCAAAUCAAGGUUU---UUAUAAAAAAUUUGUAAACAAACAAAAGCUCGACCAAAUUCAAUCUUAUCGAGCUUCAAUAUCAGCAAA ((....(((((((.((..(((((...((((.........)))))))))---..))...)))))))...........((((((((...............)))))))).....))...... ( -15.56) >DroSim_CAF1 12050 116 + 1 GAAACUCAAAUUUCAUUUGAACCAAAUUGAAGUCCCAAAUCAAGGUUU---UUAUAAAUAAUUUGUAAACAAACAAACGCUCGACCA-AUUCAAUCUUAUCGAGCUUCAAUAUCAGCAAA ((...((((((...)))))).....((((((.............((((---((((((.....))))))..))))....((((((...-...........)))))))))))).))...... ( -14.94) >DroEre_CAF1 11987 114 + 1 GCAACUCAAAUUUCAUUUAAACCGAAUUGAAGUCUCA-AUCAAGGUUUUUUUUAUAAAUAAUUUGUAAACAA----ACGCUCGACCA-AUUCCAUCUUAUCGAGCUUCAAUAUCAGCAUA ((....((((((......(((((..(((((....)))-))...)))))...........)))))).......----..((((((...-...........))))))..........))... ( -17.67) >DroYak_CAF1 13241 111 + 1 GCAACUCAAAUUUCAUUUAAACCGAAUUGAAGUCUCA-AUCAAGGUUU---UUAUAAAUAAUUUGUAAACAA----ACGCUCGACCA-AUUCAAUCUUAUCGAGCUUCAAUAUCAGCAAA ((....((((((..((..(((((..(((((....)))-))...)))))---..))....)))))).......----..((((((...-...........))))))..........))... ( -19.54) >consensus GAAACUCAAAUUUCAUUUGAACCAAAUUGAAGUCCCA_AUCAAGGUUU___UUAUAAAUAAUUUGUAAACAA____ACGCUCGACCA_AUUCAAUCUUAUCGAGCUUCAAUAUCAGCAAA .........................((((((....................((((((.....))))))..........((((((...............))))))))))))......... (-12.22 = -11.90 + -0.32)

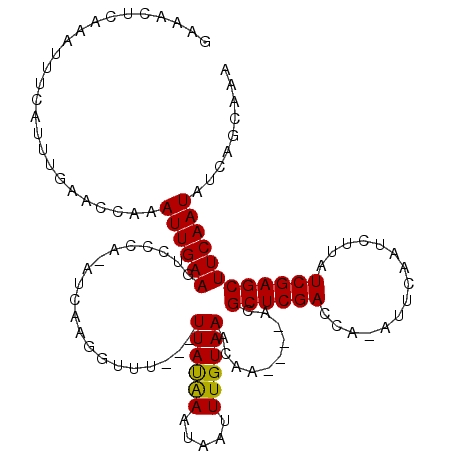
| Location | 15,588,790 – 15,588,902 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.53 |
| Mean single sequence MFE | -16.80 |
| Consensus MFE | -15.30 |
| Energy contribution | -15.14 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15588790 112 + 23771897 CAAGGUUU---UUACGAAUAAUUUGUAAACAA----ACGCUCGACCA-AUUCAAUCUUAUCGAGCUUCAAUAUCAGCAAAACUUGAUAUUUAUAAACAAAUGCGCACACCGAUUCGAAUU ...(((..---...((....((((((......----..((((((...-...........))))))...((((((((......)))))))).....)))))).))...))).......... ( -18.74) >DroSec_CAF1 12137 117 + 1 CAAGGUUU---UUAUAAAAAAUUUGUAAACAAACAAAAGCUCGACCAAAUUCAAUCUUAUCGAGCUUCAAUAUCAGCAAAACUUGAUAUUUAUAAACAAAUGCGCACACCGAUUCGAAUU ...(((..---.........((((((..........((((((((...............)))))))).((((((((......)))))))).....))))))......))).......... ( -17.59) >DroSim_CAF1 12090 116 + 1 CAAGGUUU---UUAUAAAUAAUUUGUAAACAAACAAACGCUCGACCA-AUUCAAUCUUAUCGAGCUUCAAUAUCAGCAAAACUUGAUAUUUAUAAACAAAUGCGCACACCGAUUCGAAUU ...(((..---.........((((((............((((((...-...........))))))...((((((((......)))))))).....))))))......))).......... ( -16.27) >DroEre_CAF1 12026 112 + 1 CAAGGUUUUUUUUAUAAAUAAUUUGUAAACAA----ACGCUCGACCA-AUUCCAUCUUAUCGAGCUUCAAUAUCAGCAUAACUUGAUAUUUAUAAACAAAUGCGCA---UGAUUCGAAUU ...(((((...((((.....((((((......----..((((((...-...........))))))...((((((((......)))))))).....))))))....)---)))...))))) ( -16.14) >DroYak_CAF1 13280 112 + 1 CAAGGUUU---UUAUAAAUAAUUUGUAAACAA----ACGCUCGACCA-AUUCAAUCUUAUCGAGCUUCAAUAUCAGCAAAACUUGAUAUUUAUAAACAAGUGCGCACAACGAUUCGAAUU ........---.........((((((......----..((((((...-...........))))))...((((((((......)))))))).....))))))................... ( -15.24) >consensus CAAGGUUU___UUAUAAAUAAUUUGUAAACAA____ACGCUCGACCA_AUUCAAUCUUAUCGAGCUUCAAUAUCAGCAAAACUUGAUAUUUAUAAACAAAUGCGCACACCGAUUCGAAUU ....................((((((............((((((...............))))))...((((((((......)))))))).....))))))................... (-15.30 = -15.14 + -0.16)

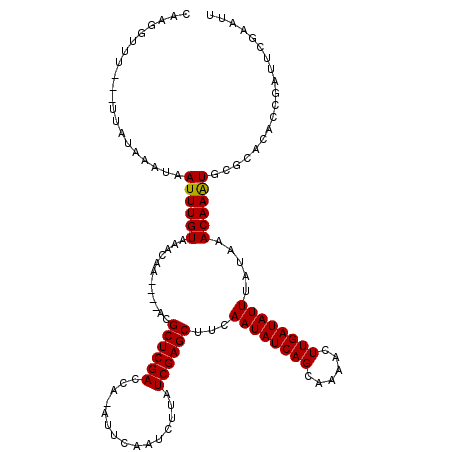
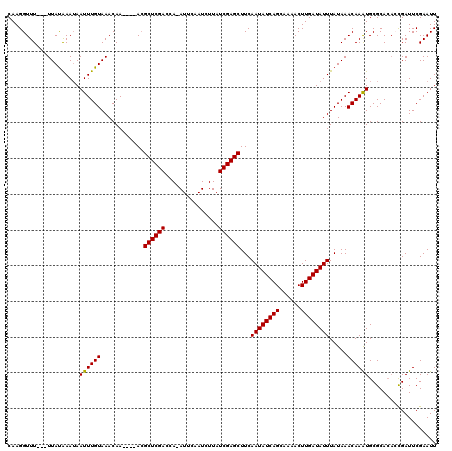
| Location | 15,588,902 – 15,589,010 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 91.24 |
| Mean single sequence MFE | -15.74 |
| Consensus MFE | -13.57 |
| Energy contribution | -14.70 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
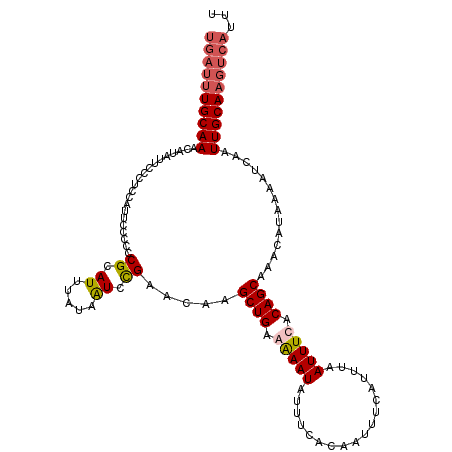
>3L_DroMel_CAF1 15588902 108 + 23771897 UGAUUUGCAAACAUAUUCCUUCCAUUCCCCCCGCAUUUAUAAUCCGAACAAGCUGAAGAAUAUUUCACAAUUCCAUUUAAUUUCACAGCAAACAUAAAAUCAAUUGCA-------- ((((((((........................)).................((((..((((..................))))..)))).......))))))......-------- ( -7.23) >DroSec_CAF1 12254 116 + 1 UGAUUUGCAAACAUAUUCCCUCCAUUCCCCCCGCAUUUAUAAUCCGAACAAGCUGAGAAAUAUUUCACAAUUUCAUUUAAUUUCACAGCAAACAUAAAAUCAAUUGCAAGUCAUUC ((((((((((.....................((.((.....)).)).....((((.(((((..................))))).))))..............))))))))))... ( -18.27) >DroSim_CAF1 12206 116 + 1 UGAUUUGCAAACAUAUUCCCUCCAUUCCCCCCGCAUUUAUAGUCCGAACAAGCUGAGAAAUAUUUCACAAUUUCAUUUAAUUUCACAGCAAACAUAAAAUCAAUUGCAAGUCAUUU ((((((((((.....................((.((.....)).)).....((((.(((((..................))))).))))..............))))))))))... ( -18.87) >DroEre_CAF1 12138 116 + 1 UGAUUUGCAAACAUAUUCCCUGCAUUCCCCCCGCAUUUAUAAUUUGAUCAAGCUGAAGAAUAUUUCACAAUUUCAUUUGAUUUCACAGCAAACAUAAAUUCAAUUGCAAGUCAUUU ((((((((((..........(((.........)))................((((..((.....)).(((......)))......))))..............))))))))))... ( -18.60) >consensus UGAUUUGCAAACAUAUUCCCUCCAUUCCCCCCGCAUUUAUAAUCCGAACAAGCUGAAAAAUAUUUCACAAUUUCAUUUAAUUUCACAGCAAACAUAAAAUCAAUUGCAAGUCAUUU ((((((((((.....................((.((.....)).)).....((((.(((((..................))))).))))..............))))))))))... (-13.57 = -14.70 + 1.13)

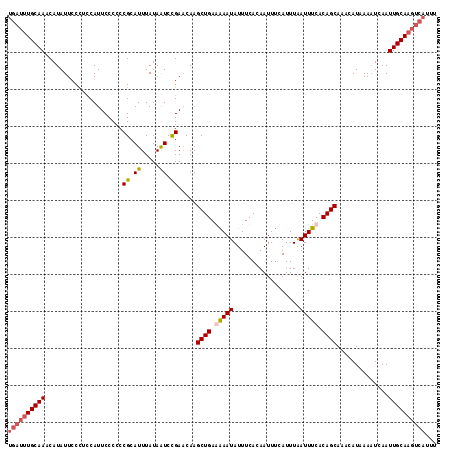
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:55 2006