
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,584,064 – 15,584,203 |
| Length | 139 |
| Max. P | 0.992756 |

| Location | 15,584,064 – 15,584,165 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 88.34 |
| Mean single sequence MFE | -23.27 |
| Consensus MFE | -19.47 |
| Energy contribution | -19.05 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15584064 101 + 23771897 ---UUUUUUUUUCAAUUUGAAUAAC---UGCUGUUGGACCAUUUGAUUUAUGUACAUUGAACUUGCUGAAAGUUUAGCUAAUACAUAAAAUGCAAACGGCGUGCGUA ---......................---.((((((....(((((....((((((..((((((((.....))))))))....)))))))))))..))))))....... ( -17.90) >DroSec_CAF1 7379 104 + 1 UUAUUUUUUUUUCAAUUUUAGCAAC---UGCUGUUGGACCAUUUGAUUUAUGUACAUUGAACUUGCUGAAAGUUCAACUAAUACAUAAAAUGCAAACGGCGUGCGUA ....................(((..---.((((((....(((((....((((((..((((((((.....))))))))....)))))))))))..)))))).)))... ( -23.80) >DroSim_CAF1 7353 101 + 1 ---UUUUUUUUUCAAUUUUAGCAAC---UGCUGUUGGACCAUUUGAUUUAUGUACAUUGAACUUGCUGAAAGUUCAACUAAUACAUAAAAUGCAAACGGCGUGCGUA ---.................(((..---.((((((....(((((....((((((..((((((((.....))))))))....)))))))))))..)))))).)))... ( -23.80) >DroEre_CAF1 7371 102 + 1 UGAUU--UUUUUUAAUUUGAGUAAC---UGCUGCUGGGCCAUUUGAUUUAUGUACAUUGAACUUGCCAAAAGUUCAACUAAUACAUAAAAUGCAAACGGCGUGCGUA .....--............(((...---.)))((.(.(((.((((.((((((((..((((((((.....))))))))....))))))))...)))).))).)))... ( -23.70) >DroYak_CAF1 8574 101 + 1 UUAUC--UUUUUU-AUUUGAGUAAC---UGCUGUUGGGCCAUUUGAUUUAUGUACAUUGAACUUGCUGAAAGUUCAACUAAUACAUAAAAUGCAAACGGCGUGCGUA .....--......-......(((..---.((((((..((.((((....((((((..((((((((.....))))))))....)))))))))))).)))))).)))... ( -22.80) >DroAna_CAF1 7703 107 + 1 UUAUAUGCCAUUUAAUUUAAGGCACUCCUGCUGCUCGUCCCUUUGAUUUAUGUACAUUGAACUUGCUGAAAGUUCAGCUAAUACAUAAAAUGCAAACGGCGUGCGUA .....((((...........))))........((.((.((.((((.((((((((..((((((((.....))))))))....))))))))...)))).)))).))... ( -27.60) >consensus UUAUUUUUUUUUCAAUUUGAGUAAC___UGCUGUUGGACCAUUUGAUUUAUGUACAUUGAACUUGCUGAAAGUUCAACUAAUACAUAAAAUGCAAACGGCGUGCGUA .............................(((((((.....((((.((((((((..((((((((.....))))))))....))))))))...))))))))).))... (-19.47 = -19.05 + -0.42)

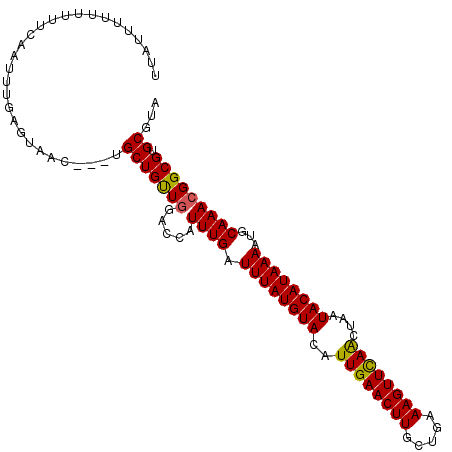

| Location | 15,584,064 – 15,584,165 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 88.34 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -22.43 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.63 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15584064 101 - 23771897 UACGCACGCCGUUUGCAUUUUAUGUAUUAGCUAAACUUUCAGCAAGUUCAAUGUACAUAAAUCAAAUGGUCCAACAGCA---GUUAUUCAAAUUGAAAAAAAAA--- .......((((((((...(((((((((......(((((.....)))))....))))))))).)))))))).......((---(((.....))))).........--- ( -18.50) >DroSec_CAF1 7379 104 - 1 UACGCACGCCGUUUGCAUUUUAUGUAUUAGUUGAACUUUCAGCAAGUUCAAUGUACAUAAAUCAAAUGGUCCAACAGCA---GUUGCUAAAAUUGAAAAAAAAAUAA .......((((((((...(((((((((..(((((((((.....)))))))))))))))))).)))))))).....(((.---...)))................... ( -26.70) >DroSim_CAF1 7353 101 - 1 UACGCACGCCGUUUGCAUUUUAUGUAUUAGUUGAACUUUCAGCAAGUUCAAUGUACAUAAAUCAAAUGGUCCAACAGCA---GUUGCUAAAAUUGAAAAAAAAA--- .......((((((((...(((((((((..(((((((((.....)))))))))))))))))).)))))))).....(((.---...)))................--- ( -26.70) >DroEre_CAF1 7371 102 - 1 UACGCACGCCGUUUGCAUUUUAUGUAUUAGUUGAACUUUUGGCAAGUUCAAUGUACAUAAAUCAAAUGGCCCAGCAGCA---GUUACUCAAAUUAAAAAA--AAUCA ...((..((((((((...(((((((((..(((((((((.....)))))))))))))))))).))))))))...))....---..................--..... ( -28.60) >DroYak_CAF1 8574 101 - 1 UACGCACGCCGUUUGCAUUUUAUGUAUUAGUUGAACUUUCAGCAAGUUCAAUGUACAUAAAUCAAAUGGCCCAACAGCA---GUUACUCAAAU-AAAAAA--GAUAA .......((((((((...(((((((((..(((((((((.....)))))))))))))))))).)))))))).........---...........-......--..... ( -26.60) >DroAna_CAF1 7703 107 - 1 UACGCACGCCGUUUGCAUUUUAUGUAUUAGCUGAACUUUCAGCAAGUUCAAUGUACAUAAAUCAAAGGGACGAGCAGCAGGAGUGCCUUAAAUUAAAUGGCAUAUAA ...((.((((.((((...(((((((((....(((((((.....)))))))..))))))))).)))).)).)).)).......(((((...........))))).... ( -31.20) >consensus UACGCACGCCGUUUGCAUUUUAUGUAUUAGUUGAACUUUCAGCAAGUUCAAUGUACAUAAAUCAAAUGGUCCAACAGCA___GUUACUCAAAUUAAAAAAAAAAUAA ...((..((((((((...(((((((((..(((((((((.....)))))))))))))))))).))))))))......))............................. (-22.43 = -23.10 + 0.67)



| Location | 15,584,086 – 15,584,203 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.09 |
| Mean single sequence MFE | -22.33 |
| Consensus MFE | -12.38 |
| Energy contribution | -13.93 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15584086 117 - 23771897 CAUUUUAUAAAUUACA-CUAACUUUUUUCUUCC-CCUGCCUACGCACGCCGUUUGCAUUUUAUGUAUUAGCUAAACUUUCAGCAAGUUCAAUGUACAUAAAUCAAAUGGUCCAACAGCA ................-................-..(((....))).((((((((...(((((((((......(((((.....)))))....))))))))).))))))))......... ( -18.20) >DroPse_CAF1 45128 102 - 1 CAUUUUAUAAAUGACUUCUUUGUUUUUUUUUGA-UUUGUUAACGCACGCUGUUUGCAUUUAAUGUAUUAGUUGAACUUUGAGCAAGUUCAAUGUUCA----------------ACGGAA ((..(((.(((.(((......)))..))).)))-..)).........((.....)).............(((((((.(((((....))))).)))))----------------)).... ( -17.30) >DroEre_CAF1 7394 117 - 1 CAUUUUAUAAAUUACA-CUAACUUUUUUCUUCC-CUUGCCUACGCACGCCGUUUGCAUUUUAUGUAUUAGUUGAACUUUUGGCAAGUUCAAUGUACAUAAAUCAAAUGGCCCAGCAGCA ................-................-.........((..((((((((...(((((((((..(((((((((.....)))))))))))))))))).))))))))...)).... ( -28.60) >DroYak_CAF1 8596 116 - 1 CAUUUUAUAAAUUACA-CUAACUUUUUUCUUCC-C-UGCCUACGCACGCCGUUUGCAUUUUAUGUAUUAGUUGAACUUUCAGCAAGUUCAAUGUACAUAAAUCAAAUGGCCCAACAGCA ................-................-.-(((....))).((((((((...(((((((((..(((((((((.....)))))))))))))))))).))))))))......... ( -27.50) >DroAna_CAF1 7731 118 - 1 CAUUUUAUAAAUUACA-CUAACUUUUUUCCUCGAUUUGUCUACGCACGCCGUUUGCAUUUUAUGUAUUAGCUGAACUUUCAGCAAGUUCAAUGUACAUAAAUCAAAGGGACGAGCAGCA ................-..........................((.((((.((((...(((((((((....(((((((.....)))))))..))))))))).)))).)).)).)).... ( -25.10) >DroPer_CAF1 45313 102 - 1 CAUUUUAUAAAUGACUUCUUUGUUUUUUUUUGA-UUUGUUAACGCACGCUGUUUGCAUUUAAUGUAUUAGUUGAACUUUGAGCAAGUUCAACGUUCA----------------ACGGAA ((..(((.(((.(((......)))..))).)))-..)).........((.....)).............(((((((.(((((....))))).)))))----------------)).... ( -17.30) >consensus CAUUUUAUAAAUUACA_CUAACUUUUUUCUUCC_CUUGCCUACGCACGCCGUUUGCAUUUUAUGUAUUAGUUGAACUUUCAGCAAGUUCAAUGUACAUAAAUCAAAUGG_CCAACAGCA ....................................(((..(((.....)))..))).(((((((((..(((((((((.....)))))))))))))))))).................. (-12.38 = -13.93 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:47 2006