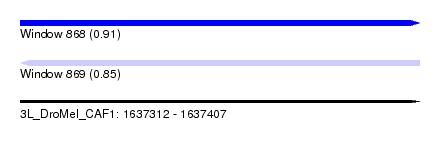
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,637,312 – 1,637,407 |
| Length | 95 |
| Max. P | 0.914129 |

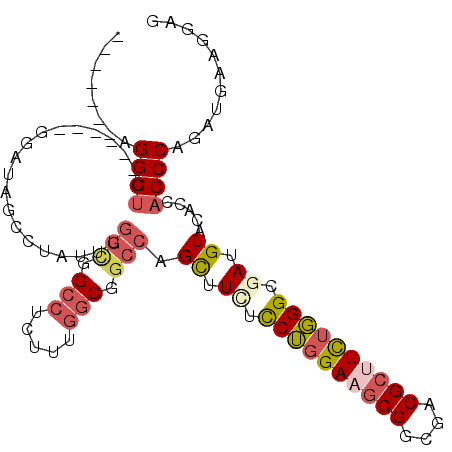
| Location | 1,637,312 – 1,637,407 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.66 |
| Mean single sequence MFE | -34.22 |
| Consensus MFE | -24.32 |
| Energy contribution | -25.35 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1637312 95 + 23771897 CUCCUUCAUCUGGGUGGUGUACAUCGCCCAGAAGCGUCGCCGCUUCCAGGACAAGCUGGCCGCCAAAGAGGCAGCCAAAUAGGCUAUAC------GACCCU------- ........((((((((((....))))))))))..(((.(((((((.......))))((((.(((.....))).))))....)))...))------).....------- ( -36.20) >DroSim_CAF1 8233 95 + 1 CUCCUUCAUCUGGGUGGUAUACAUCGCCCAAAGGCGUCGCCGCUUCCAGGAAAAGCUGGCCGCCAAAGAGGCAGCCAAAUAGGCUAUCU------GACCCU------- ..........((((((((....)))))))).(((.((((((((((.......))))((((.(((.....))).))))....))).....------))))))------- ( -33.50) >DroEre_CAF1 8243 95 + 1 CUCCUUCAUCUGGGUGGUGUACAUCGCCCAGAGGCGUCGCCGCUUCCAGGAAAAGCAGGCAGCCAAAGAGGCCGCCAAAUAGGCUAUCC------GACCCU------- ........((((((((((....))))))))))((.((((((((((.......)))).(((.(((.....))).))).....))).....------))))).------- ( -38.50) >DroYak_CAF1 8209 95 + 1 CUCCUUCAUCUGGGUGGUGUACAUCGCCCAGAAGCGUCGCCGAUUCCAGGAGAAGCAGGCUGCCAAAAAGGCCGCCAAAUAGGCCAUCC------GACCCU------- ........((((((((((....))))))))))((.((((((..(((.....)))...))).........((((........))))....------))).))------- ( -34.20) >DroAna_CAF1 8957 102 + 1 UUCCUUCAUCUGGGUGGUCUACAUCGCCCAGAAGCGCCGCCGCUUCCAGGAGAAACUGGCCGCCGAAAAGGCCACCAAAUAAGCAUUCC------GUCCCCAUCCGGA ........(((((((((...((.((.((..((((((....))))))..)).))...(((((........)))))...............------))..))))))))) ( -34.30) >DroPer_CAF1 8704 98 + 1 UUCCUUCAUUUGGGUAGUUUACAUUGCCAAGAAGCGUCGUCGCUUCGCGAAUAAGCAGGCUGCUGACAAGUC---GAAAUAAACAUUCCAGUGGGGACCCC------- .((((.((((.(((..(((((....(((..((((((....))))))((......)).))).(((....))).---....)))))..)))))))))))....------- ( -28.60) >consensus CUCCUUCAUCUGGGUGGUGUACAUCGCCCAGAAGCGUCGCCGCUUCCAGGAAAAGCAGGCCGCCAAAAAGGCCGCCAAAUAGGCUAUCC______GACCCU_______ .((((.....((((((((....))))))))((((((....)))))).))))......(((.(((.....))).)))................................ (-24.32 = -25.35 + 1.03)

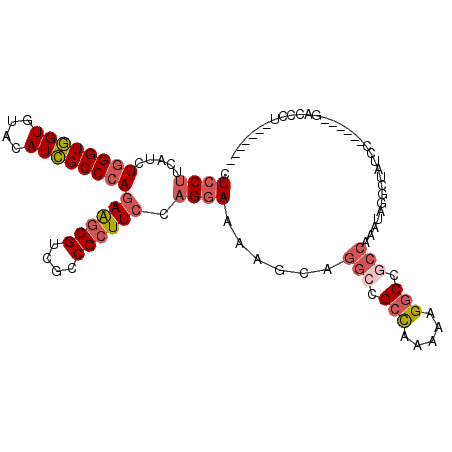
| Location | 1,637,312 – 1,637,407 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.66 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -26.51 |
| Energy contribution | -27.60 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1637312 95 - 23771897 -------AGGGUC------GUAUAGCCUAUUUGGCUGCCUCUUUGGCGGCCAGCUUGUCCUGGAAGCGGCGACGCUUCUGGGCGAUGUACACCACCCAGAUGAAGGAG -------.((((.------(((((......(((((((((.....))))))))).(((.((..((((((....))))))..))))))))))...))))........... ( -44.90) >DroSim_CAF1 8233 95 - 1 -------AGGGUC------AGAUAGCCUAUUUGGCUGCCUCUUUGGCGGCCAGCUUUUCCUGGAAGCGGCGACGCCUUUGGGCGAUGUAUACCACCCAGAUGAAGGAG -------.((((.------..((((((....((((((((.....))))))))(((((.....))))))))..((((....))))...)))...))))........... ( -39.60) >DroEre_CAF1 8243 95 - 1 -------AGGGUC------GGAUAGCCUAUUUGGCGGCCUCUUUGGCUGCCUGCUUUUCCUGGAAGCGGCGACGCCUCUGGGCGAUGUACACCACCCAGAUGAAGGAG -------.((((.------((...(((.....(((((((.....))))))).(((((.....))))))))..((((....)))).......))))))........... ( -40.60) >DroYak_CAF1 8209 95 - 1 -------AGGGUC------GGAUGGCCUAUUUGGCGGCCUUUUUGGCAGCCUGCUUCUCCUGGAAUCGGCGACGCUUCUGGGCGAUGUACACCACCCAGAUGAAGGAG -------.((((.------((.((........(((.(((.....))).)))(((.((.((..(((.((....)).)))..)).)).)))))))))))........... ( -35.90) >DroAna_CAF1 8957 102 - 1 UCCGGAUGGGGAC------GGAAUGCUUAUUUGGUGGCCUUUUCGGCGGCCAGUUUCUCCUGGAAGCGGCGGCGCUUCUGGGCGAUGUAGACCACCCAGAUGAAGGAA (((...((((...------((..(((....(((((.(((.....))).)))))..((.((..((((((....))))))..)).)).)))..)).))))......))). ( -42.70) >DroPer_CAF1 8704 98 - 1 -------GGGGUCCCCACUGGAAUGUUUAUUUC---GACUUGUCAGCAGCCUGCUUAUUCGCGAAGCGACGACGCUUCUUGGCAAUGUAAACUACCCAAAUGAAGGAA -------(((.(((.....)))..((((((...---((....))....(((.((......))((((((....))))))..)))...))))))..)))........... ( -24.80) >consensus _______AGGGUC______GGAUAGCCUAUUUGGCGGCCUCUUUGGCGGCCAGCUUCUCCUGGAAGCGGCGACGCUUCUGGGCGAUGUACACCACCCAGAUGAAGGAG ........((((....................(((.(((.....))).))).((.((.((((((((((....)))))))))).)).)).....))))........... (-26.51 = -27.60 + 1.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:32 2006