
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,570,773 – 15,570,905 |
| Length | 132 |
| Max. P | 0.996123 |

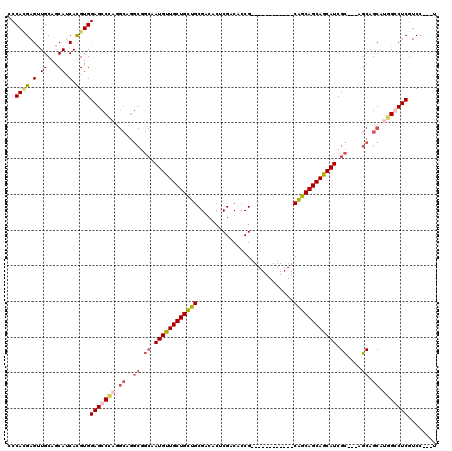
| Location | 15,570,773 – 15,570,875 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.03 |
| Mean single sequence MFE | -49.95 |
| Consensus MFE | -26.70 |
| Energy contribution | -28.33 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15570773 102 + 23771897 CCCACGAGUUGCAGCAUCAUGUGGAGCCCAGGCAGGCGGCCAUGUUGCUGCUGCGACACUCGACACCG------------CAGCAGCAACAUCGC---AGCAGCAUGGCCUCGUCC---U ...((((((..((......))..).(((...((..((.((.((((((((((((((...........))------------)))))))))))).))---.)).))..))))))))..---. ( -45.40) >DroEre_CAF1 33476 102 + 1 CCCAAGAGUUGCAGCAUCACGUGGAGCCCAGGCAGGCGGCAAUGUUGCUGCUGCGGCACUCGACGCCG------------CAGCAGCAGCAUCGC---AGCAGCAUGGGCUCGUCC---U .....((((....)).)).....(((((((.((..((.((.((((((((((((((((.......))))------------)))))))))))).))---.)).)).)))))))....---. ( -56.10) >DroYak_CAF1 53103 102 + 1 CCCACGAGUUGCAGCAUCACGUGGAGCCCAGGCAGGCGGCAAUGUUGCUGCUGCGCCACUCGACGCCG------------CAGCAGCAGCAUCGC---AGCAGCACUGGCUCGUCC---U ..((((.(.((...)).).))))(((((.((((..((.((.((((((((((((((.(.......).))------------)))))))))))).))---.)).)).)))))))....---. ( -47.80) >DroAna_CAF1 33016 111 + 1 CCCAUGAGCUGCAACAGCAUGUGGAGCCGAG---------CAUGUUGCUGGUGCGUCACUCGACACCGAUGCUGGUGGCGCGUCAGCAGCAUCGGCUGGCCAGCAAUGCCUCCUCCUCGU .(((((.((((...)))).)))))((((((.---------..((((((((..((((((((.(.((....))).))))))))..))))))))))))))(((.(....)))).......... ( -50.50) >consensus CCCACGAGUUGCAGCAUCACGUGGAGCCCAGGCAGGCGGCAAUGUUGCUGCUGCGACACUCGACACCG____________CAGCAGCAGCAUCGC___AGCAGCAUGGCCUCGUCC___U ..((((.(.((...)).).))))(((((((.((..((.((.((((((((((((...........................)))))))))))).))....)).)).)))))))........ (-26.70 = -28.33 + 1.63)


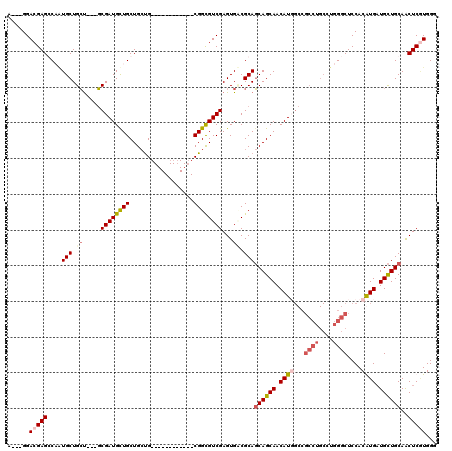
| Location | 15,570,773 – 15,570,875 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.03 |
| Mean single sequence MFE | -51.80 |
| Consensus MFE | -26.96 |
| Energy contribution | -29.02 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15570773 102 - 23771897 A---GGACGAGGCCAUGCUGCU---GCGAUGUUGCUGCUG------------CGGUGUCGAGUGUCGCAGCAGCAACAUGGCCGCCUGCCUGGGCUCCACAUGAUGCUGCAACUCGUGGG .---....(((.(((.((.((.---((.((((((((((((------------(((..(...)..))))))))))))))).)).))..)).))).)))..(((((.(......)))))).. ( -51.40) >DroEre_CAF1 33476 102 - 1 A---GGACGAGCCCAUGCUGCU---GCGAUGCUGCUGCUG------------CGGCGUCGAGUGCCGCAGCAGCAACAUUGCCGCCUGCCUGGGCUCCACGUGAUGCUGCAACUCUUGGG (---(((.(((((((.((.((.---((((((.((((((((------------(((((.....))))))))))))).)))))).))..)).)))))))...((......))...))))... ( -56.50) >DroYak_CAF1 53103 102 - 1 A---GGACGAGCCAGUGCUGCU---GCGAUGCUGCUGCUG------------CGGCGUCGAGUGGCGCAGCAGCAACAUUGCCGCCUGCCUGGGCUCCACGUGAUGCUGCAACUCGUGGG .---..(((((...((((..((---.(((((((((....)------------))))))))))..)))).((((((.(((((..((((....))))..)).))).))))))..)))))... ( -53.60) >DroAna_CAF1 33016 111 - 1 ACGAGGAGGAGGCAUUGCUGGCCAGCCGAUGCUGCUGACGCGCCACCAGCAUCGGUGUCGAGUGACGCACCAGCAACAUG---------CUCGGCUCCACAUGCUGUUGCAGCUCAUGGG ....((((..(((((((((((...((((((((((.((......)).))))))))))(((....)))...))))))..)))---------).)..)))).(((((((...))))..))).. ( -45.70) >consensus A___GGACGAGCCAAUGCUGCU___GCGAUGCUGCUGCUG____________CGGCGUCGAGUGACGCAGCAGCAACAUGGCCGCCUGCCUGGGCUCCACAUGAUGCUGCAACUCGUGGG ......(((((....(((.((.....((((((((..................))))))))...)).)))((((((.((((...((((....))))....)))).))))))..)))))... (-26.96 = -29.02 + 2.06)

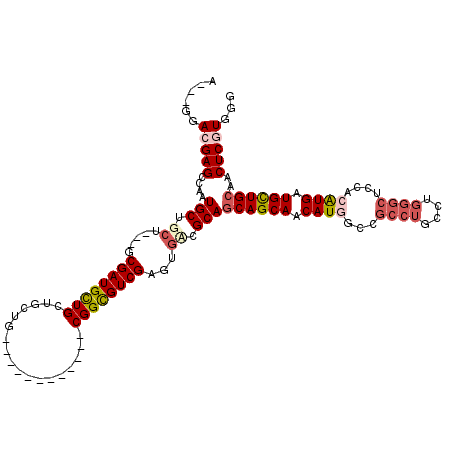
| Location | 15,570,813 – 15,570,905 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.42 |
| Mean single sequence MFE | -38.52 |
| Consensus MFE | -19.99 |
| Energy contribution | -19.24 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15570813 92 + 23771897 CAUGUUGCUGCUGCGACACUCGACACCG------------CAGCAGCAACAUCGC---AGCAGCAUGGCCUCGUCC---UUGUACCAAUCCUUUGACGUGACCUCCGCCG---------- .((((((((((((((...........))------------)))))))))))).((---....))..(((...(((.---.(((..(((....)))))).)))....))).---------- ( -32.50) >DroEre_CAF1 33516 92 + 1 AAUGUUGCUGCUGCGGCACUCGACGCCG------------CAGCAGCAGCAUCGC---AGCAGCAUGGGCUCGUCC---UUGUACCAAUCCUUUGACGUGACCUCCGCCG---------- .((((((((((((((((.......))))------------))))))))))))...---....((..((((.((((.---..(........)...)))).).)))..))..---------- ( -40.30) >DroYak_CAF1 53143 92 + 1 AAUGUUGCUGCUGCGCCACUCGACGCCG------------CAGCAGCAGCAUCGC---AGCAGCACUGGCUCGUCC---UUGUACCAAUCCUUUGACGUGACCUCCGCCG---------- .((((((((((((((.(.......).))------------)))))))))))).((---....))...(((..(((.---.(((..(((....)))))).)))....))).---------- ( -32.50) >DroAna_CAF1 33047 120 + 1 CAUGUUGCUGGUGCGUCACUCGACACCGAUGCUGGUGGCGCGUCAGCAGCAUCGGCUGGCCAGCAAUGCCUCCUCCUCGUUGUACCAAUCGUUCGAGGUGACCUCCACCACGCUGCCGAU ..((((((((..((((((((.(.((....))).))))))))..))))))))(((((.(((..((((((.........)))))).............((((.....))))..)))))))). ( -48.80) >consensus AAUGUUGCUGCUGCGACACUCGACACCG____________CAGCAGCAGCAUCGC___AGCAGCAUGGCCUCGUCC___UUGUACCAAUCCUUUGACGUGACCUCCGCCG__________ .((((((((((((...........................)))))))))))).((....)).((..((.(.((((...................)))).).))...))............ (-19.99 = -19.24 + -0.75)

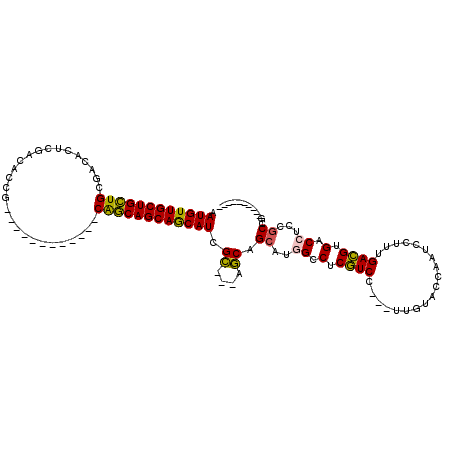
| Location | 15,570,813 – 15,570,905 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.42 |
| Mean single sequence MFE | -43.60 |
| Consensus MFE | -22.07 |
| Energy contribution | -23.26 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15570813 92 - 23771897 ----------CGGCGGAGGUCACGUCAAAGGAUUGGUACAA---GGACGAGGCCAUGCUGCU---GCGAUGUUGCUGCUG------------CGGUGUCGAGUGUCGCAGCAGCAACAUG ----------((((((.((((.((((...(........)..---.)))).))))...)))))---)..((((((((((((------------(((..(...)..))))))))))))))). ( -46.40) >DroEre_CAF1 33516 92 - 1 ----------CGGCGGAGGUCACGUCAAAGGAUUGGUACAA---GGACGAGCCCAUGCUGCU---GCGAUGCUGCUGCUG------------CGGCGUCGAGUGCCGCAGCAGCAACAUU ----------((((((.((.(.((((...(........)..---.)))).).))...)))))---).((((.((((((((------------(((((.....))))))))))))).)))) ( -43.80) >DroYak_CAF1 53143 92 - 1 ----------CGGCGGAGGUCACGUCAAAGGAUUGGUACAA---GGACGAGCCAGUGCUGCU---GCGAUGCUGCUGCUG------------CGGCGUCGAGUGGCGCAGCAGCAACAUU ----------.((((.......)))).....((((((.(..---....).))))))((((((---((((((((((....)------------)))))))(.....)))))))))...... ( -39.70) >DroAna_CAF1 33047 120 - 1 AUCGGCAGCGUGGUGGAGGUCACCUCGAACGAUUGGUACAACGAGGAGGAGGCAUUGCUGGCCAGCCGAUGCUGCUGACGCGCCACCAGCAUCGGUGUCGAGUGACGCACCAGCAACAUG .......((.(((((...(((((.((((.....((((.((.(((.(......).))).))))))((((((((((.((......)).)))))))))).))))))))).)))))))...... ( -44.50) >consensus __________CGGCGGAGGUCACGUCAAAGGAUUGGUACAA___GGACGAGCCAAUGCUGCU___GCGAUGCUGCUGCUG____________CGGCGUCGAGUGACGCAGCAGCAACAUG ...........(((((.((((.((((......(((...)))....)))).))))...)))))......(((.(((((((...............(((((....)))))))))))).))). (-22.07 = -23.26 + 1.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:39 2006