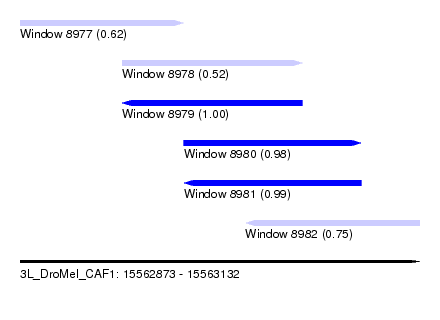
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,562,873 – 15,563,132 |
| Length | 259 |
| Max. P | 0.999076 |

| Location | 15,562,873 – 15,562,979 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.30 |
| Mean single sequence MFE | -13.50 |
| Consensus MFE | -7.47 |
| Energy contribution | -7.51 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15562873 106 + 23771897 AAAUGGUCCGAUAUAUUUCAUUUUUAUGAAUGAAAAAAAAAAA---------AAGAAAAAACCAGCAAAGCAAGCAGCCAACGGCAUGGCAAAUAAACAAAGUUAACAAUAAAUG ...............((((((((....))))))))........---------............((.......)).((((......))))......................... ( -11.90) >DroSec_CAF1 25314 97 + 1 AAAAUCUCCGACAUUUUUCAUUUUUAUGAAUGAAAAAAA-AAA---------AAGGAAAAACCAGCAAAGCAC--------AGGCAGGGCAAAUAAACAAAGUUAACAAUAAAUG ......(((....((((((((((....))))))))))..-...---------..))).......((...((..--------..))...))......................... ( -12.60) >DroSim_CAF1 25791 95 + 1 AAAAUCUCCGACAUUUUUCAUUUUUAUGAAUGAAAAAAA-AAA---------GAGAAAAAACCAGCAAAGC----------AAGCAGGGCAAAUAAACAAAGUUAACAAUAAAUG ....((((.....((((((((((....))))))))))..-...---------)))).....((.((.....----------..)).))........................... ( -13.90) >DroEre_CAF1 25631 91 + 1 AAAUUGUCCGACAUAUUUCAUUUUUAUGAAUGAAAAAAA--------------------AGCCAGCGAAGC----AGCCAACAGCAGGGCAAAAAAACAAAGUUAACAAUAAAUG ..(((((..(((...((((((((....))))))))....--------------------.(((.((...))----.((.....))..)))...........))).)))))..... ( -15.80) >DroYak_CAF1 45828 110 + 1 AAAUUGUCCGACAUAUUUCAUUUUUAUGAAUGAAAAAAA-AAAAAAAACGAAAAGAAACAGCCAGCGAAGC----AGUCAACAGCAGGGUAAAUAAACAAAGUUAACAAUAAAUG ..(((((..(((...((((((((....))))))))....-....................(((.((.....----........))..)))...........))).)))))..... ( -13.32) >consensus AAAUUGUCCGACAUAUUUCAUUUUUAUGAAUGAAAAAAA_AAA_________AAGAAAAAACCAGCAAAGC____AG_CAACAGCAGGGCAAAUAAACAAAGUUAACAAUAAAUG .........(((...((((((((....))))))))..........................((.((.................)).)).............)))........... ( -7.47 = -7.51 + 0.04)

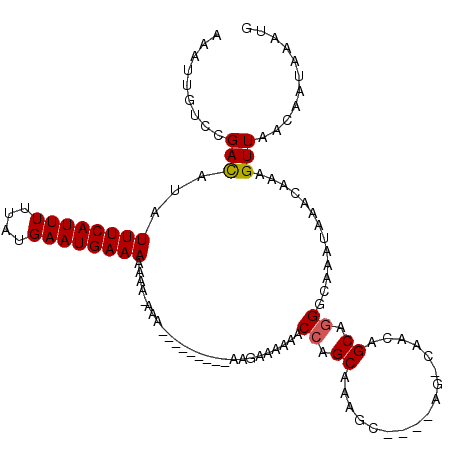
| Location | 15,562,939 – 15,563,056 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 90.70 |
| Mean single sequence MFE | -21.22 |
| Consensus MFE | -15.28 |
| Energy contribution | -14.72 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15562939 117 + 23771897 AGCCAACGGCAUGGCAAAUAAACAAAGUUAACAAUAAAUGAAAAUGAUUCAUCUGAAUAAAAUGACGAGUUUGCCGGAUUGUCAAUGAACAAUACGCAGAAGCUACGUUUGCCAAAU .(((...))).((((((((.......((((.......((....)).((((....))))....))))(..(((((...(((((......)))))..)))))..)...))))))))... ( -24.60) >DroSec_CAF1 25377 110 + 1 ------AGGCAGGGCAAAUAAACAAAGUUAACAAUAAAUGAAAAUGAUUCAUCUAAAUAAAAUGACGAGU-UGUCCGAUUGUCAAUGAACAAUACGCAGAAGCUACGCCCGCCAAAU ------.(((.((((...........(((.....((.(((((.....))))).)).......((((((..-.......))))))...))).....((....))...))))))).... ( -22.50) >DroSim_CAF1 25852 111 + 1 ------AAGCAGGGCAAAUAAACAAAGUUAACAAUAAAUGAAGAUGAUUCAUCUAAAUAAAAUGACGAGUUUGUCCGAUUGUCAAUGAACAAUACGCAGAAGCUACGCCUGCCAAAU ------..(((((............................(((((...)))))...........(((((((.(.(((((((......))))).)).).))))).)))))))..... ( -20.70) >DroEre_CAF1 25682 116 + 1 AGCCAACAGCAGGGCAAAAAAACAAAGUUAACAAUAAAUGAAAAUGAUUCAUCUAAAUAAAAUGACGAGU-UUUCUGAUUGUCAAUGAACAAUACGCAGAAGCUACGCCUGCCAAAU ........(((((.(...........(((.(((((....((((((...((((.........))))...))-))))..))))).))).........((....))...))))))..... ( -18.80) >DroYak_CAF1 45898 116 + 1 AGUCAACAGCAGGGUAAAUAAACAAAGUUAACAAUAAAUGAAAAUGAUUCAUCUAAAUAAAAUGACGAGU-UGUCUGAUUGUCAAUGAACAAUACGCAGAAGCUACGCCUGCGAAAU ........((((((((..........(((.....((.(((((.....))))).)).......((((((..-.......))))))...))).....((....))))).)))))..... ( -19.50) >consensus AG_CAACAGCAGGGCAAAUAAACAAAGUUAACAAUAAAUGAAAAUGAUUCAUCUAAAUAAAAUGACGAGU_UGUCCGAUUGUCAAUGAACAAUACGCAGAAGCUACGCCUGCCAAAU ........((.((((...........(((.....((.(((((.....))))).)).......((((((..........))))))...))).....((....))...))))))..... (-15.28 = -14.72 + -0.56)

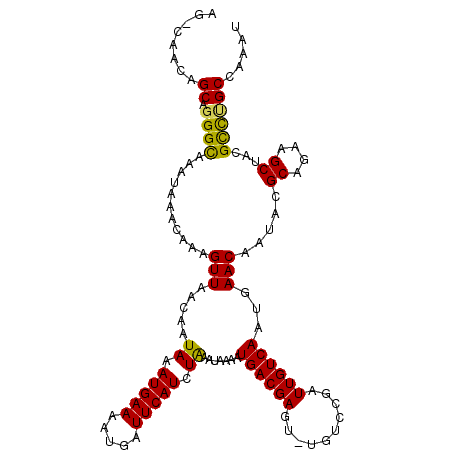
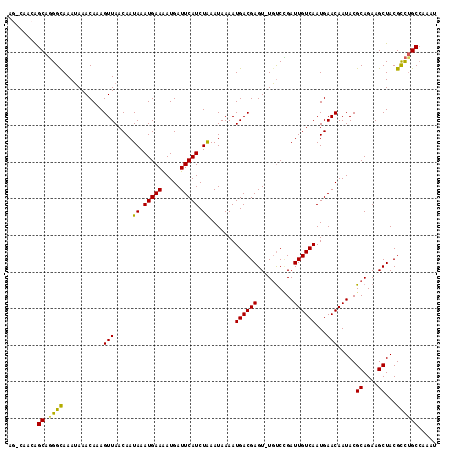
| Location | 15,562,939 – 15,563,056 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 90.70 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -23.04 |
| Energy contribution | -23.40 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.36 |
| SVM RNA-class probability | 0.999076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15562939 117 - 23771897 AUUUGGCAAACGUAGCUUCUGCGUAUUGUUCAUUGACAAUCCGGCAAACUCGUCAUUUUAUUCAGAUGAAUCAUUUUCAUUUAUUGUUAACUUUGUUUAUUUGCCAUGCCGUUGGCU ...((((((((((((...))))))........((((((((..(((......))).........(((((((.....)))))))))))))))..........)))))).(((...))). ( -26.70) >DroSec_CAF1 25377 110 - 1 AUUUGGCGGGCGUAGCUUCUGCGUAUUGUUCAUUGACAAUCGGACA-ACUCGUCAUUUUAUUUAGAUGAAUCAUUUUCAUUUAUUGUUAACUUUGUUUAUUUGCCCUGCCU------ ....(((((((((((...))))..........(((((((...(((.-....)))........((((((((.....)))))))))))))))............)))).))).------ ( -27.70) >DroSim_CAF1 25852 111 - 1 AUUUGGCAGGCGUAGCUUCUGCGUAUUGUUCAUUGACAAUCGGACAAACUCGUCAUUUUAUUUAGAUGAAUCAUCUUCAUUUAUUGUUAACUUUGUUUAUUUGCCCUGCUU------ ....((((((.(((((....))..........(((((((...(((......)))........((((((((.....)))))))))))))))...........))))))))).------ ( -25.00) >DroEre_CAF1 25682 116 - 1 AUUUGGCAGGCGUAGCUUCUGCGUAUUGUUCAUUGACAAUCAGAAA-ACUCGUCAUUUUAUUUAGAUGAAUCAUUUUCAUUUAUUGUUAACUUUGUUUUUUUGCCCUGCUGUUGGCU ....((((((.(((((....))..........((((((((..((((-(....((((((.....))))))....)))))....))))))))...........)))))))))....... ( -25.30) >DroYak_CAF1 45898 116 - 1 AUUUCGCAGGCGUAGCUUCUGCGUAUUGUUCAUUGACAAUCAGACA-ACUCGUCAUUUUAUUUAGAUGAAUCAUUUUCAUUUAUUGUUAACUUUGUUUAUUUACCCUGCUGUUGACU .....(((((.(((((....))..........(((((((...(((.-....)))........((((((((.....)))))))))))))))...........))))))))........ ( -25.40) >consensus AUUUGGCAGGCGUAGCUUCUGCGUAUUGUUCAUUGACAAUCAGACA_ACUCGUCAUUUUAUUUAGAUGAAUCAUUUUCAUUUAUUGUUAACUUUGUUUAUUUGCCCUGCUGUUG_CU ....((((((.(((((....))..........(((((((...(((......)))........((((((((.....)))))))))))))))...........)))))))))....... (-23.04 = -23.40 + 0.36)

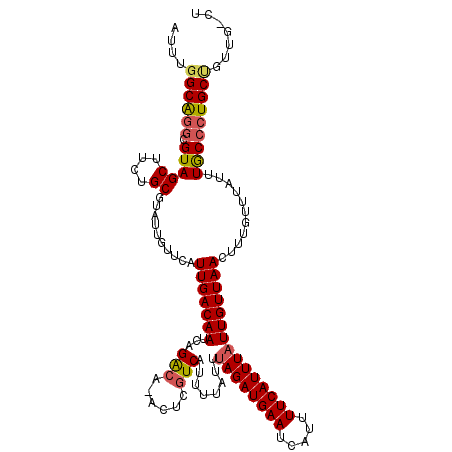
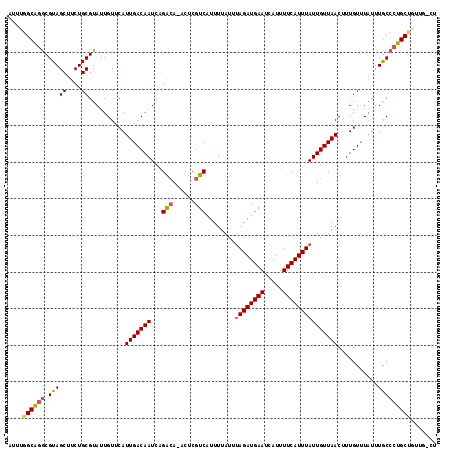
| Location | 15,562,979 – 15,563,094 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -22.94 |
| Consensus MFE | -20.98 |
| Energy contribution | -20.82 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15562979 115 + 23771897 AAAAUGAUUCAUCUGAAUAAAAUGACGAGUUUGCCGGAUUGUCAAUGAACAAUACGCAGAAGCUACGUUUGCCAAAUGCUAAAUAUUUUCAAUAUCGGAAACUAUUUAGCACUGA .(((((((((....))))........(..(((((...(((((......)))))..)))))..)..)))))......((((((((((((((......))))).))))))))).... ( -23.30) >DroSec_CAF1 25411 114 + 1 AAAAUGAUUCAUCUAAAUAAAAUGACGAGU-UGUCCGAUUGUCAAUGAACAAUACGCAGAAGCUACGCCCGCCAAAUGCUAAAUAUUUUCAAUAUCGGAAACUAUUUAGCACUGA ...(..((((.((..........)).))))-..).(((((((......))))).))(((..((.......))....((((((((((((((......))))).)))))))))))). ( -20.20) >DroSim_CAF1 25886 115 + 1 AAGAUGAUUCAUCUAAAUAAAAUGACGAGUUUGUCCGAUUGUCAAUGAACAAUACGCAGAAGCUACGCCUGCCAAAUGCUAAAUAUUUUCAAUAUCGGAAACUAUUUAGCACUGA .(((((...)))))...........(((((((.(.(((((((......))))).)).).))))).)).........((((((((((((((......))))).))))))))).... ( -23.30) >DroEre_CAF1 25722 114 + 1 AAAAUGAUUCAUCUAAAUAAAAUGACGAGU-UUUCUGAUUGUCAAUGAACAAUACGCAGAAGCUACGCCUGCCAAAUGCUAAAUAUUUUCAAAUUCGGAAACUAUUUAGCACUGG .........................(((((-((.((((((((......)))))...)))))))).))....(((..((((((((((((((......))))).))))))))).))) ( -21.10) >DroYak_CAF1 45938 114 + 1 AAAAUGAUUCAUCUAAAUAAAAUGACGAGU-UGUCUGAUUGUCAAUGAACAAUACGCAGAAGCUACGCCUGCGAAAUGCUAAAUAUUUCCAAAAUCGGAAACUAUUUAGCACUGG ....((.(((((..........((((((..-.......)))))))))))))...(((((.........)))))...((((((((((((((......))))).))))))))).... ( -26.80) >consensus AAAAUGAUUCAUCUAAAUAAAAUGACGAGU_UGUCCGAUUGUCAAUGAACAAUACGCAGAAGCUACGCCUGCCAAAUGCUAAAUAUUUUCAAUAUCGGAAACUAUUUAGCACUGA ....((.(((((..........((((((..........))))))))))))).....(((..((.......))....((((((((((((((......))))).)))))))))))). (-20.98 = -20.82 + -0.16)

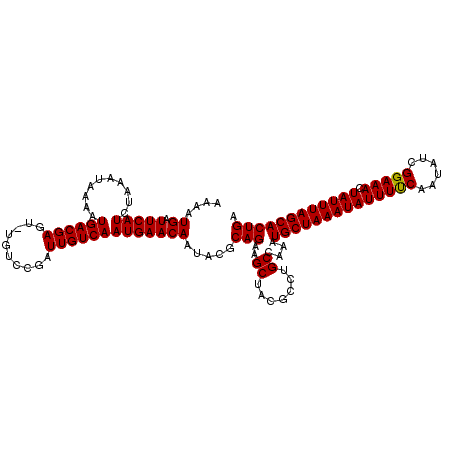
| Location | 15,562,979 – 15,563,094 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -26.79 |
| Consensus MFE | -23.45 |
| Energy contribution | -22.85 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15562979 115 - 23771897 UCAGUGCUAAAUAGUUUCCGAUAUUGAAAAUAUUUAGCAUUUGGCAAACGUAGCUUCUGCGUAUUGUUCAUUGACAAUCCGGCAAACUCGUCAUUUUAUUCAGAUGAAUCAUUUU ...((((((((((.((((.......)))).))))))))))(((.(..((((((...))))))((((((....))))))..).)))..(((((..........)))))........ ( -24.60) >DroSec_CAF1 25411 114 - 1 UCAGUGCUAAAUAGUUUCCGAUAUUGAAAAUAUUUAGCAUUUGGCGGGCGUAGCUUCUGCGUAUUGUUCAUUGACAAUCGGACA-ACUCGUCAUUUUAUUUAGAUGAAUCAUUUU (((((((((((((.((((.......)))).)))))))))).((((((((((((...)))))..((((((..........)))))-))))))))...........)))........ ( -27.70) >DroSim_CAF1 25886 115 - 1 UCAGUGCUAAAUAGUUUCCGAUAUUGAAAAUAUUUAGCAUUUGGCAGGCGUAGCUUCUGCGUAUUGUUCAUUGACAAUCGGACAAACUCGUCAUUUUAUUUAGAUGAAUCAUCUU ..(((((((((((.((((.......)))).)))))))))))((((((((((((...)))))).((((((..........)))))).)).))))........(((((...))))). ( -27.20) >DroEre_CAF1 25722 114 - 1 CCAGUGCUAAAUAGUUUCCGAAUUUGAAAAUAUUUAGCAUUUGGCAGGCGUAGCUUCUGCGUAUUGUUCAUUGACAAUCAGAAA-ACUCGUCAUUUUAUUUAGAUGAAUCAUUUU (((((((((((((.((((.......)))).))))))))).))))..((((....(((((...((((((....))))))))))).-...))))....................... ( -26.30) >DroYak_CAF1 45938 114 - 1 CCAGUGCUAAAUAGUUUCCGAUUUUGGAAAUAUUUAGCAUUUCGCAGGCGUAGCUUCUGCGUAUUGUUCAUUGACAAUCAGACA-ACUCGUCAUUUUAUUUAGAUGAAUCAUUUU ..(((((((((((.((((((....))))))))))))))))).((((((.......))))))....(((((((........(((.-....)))..........)))))))...... ( -28.17) >consensus UCAGUGCUAAAUAGUUUCCGAUAUUGAAAAUAUUUAGCAUUUGGCAGGCGUAGCUUCUGCGUAUUGUUCAUUGACAAUCAGACA_ACUCGUCAUUUUAUUUAGAUGAAUCAUUUU ..(((((((((((.((((.......)))).)))))))))))..((((((((((...)))))).))))(((((........(((......)))..........)))))........ (-23.45 = -22.85 + -0.60)

| Location | 15,563,019 – 15,563,132 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 92.40 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -20.80 |
| Energy contribution | -20.24 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15563019 113 - 23771897 ACACGGCCAAAACAUUAGCAUAAAAC--UAUUUCCACGAAUCAGUGCUAAAUAGUUUCCGAUAUUGAAAAUAUUUAGCAUUUGGCAAACGUAGCUUCUGCGUAUUGUUCAUUGAC .....((((((.............((--(..(((...)))..)))((((((((.((((.......)))).)))))))).))))))..((((((...))))))............. ( -24.10) >DroSec_CAF1 25450 115 - 1 AACCGGCCAAAACAUUAGCAUACAGCCAUAUUUCCACGAAUCAGUGCUAAAUAGUUUCCGAUAUUGAAAAUAUUUAGCAUUUGGCGGGCGUAGCUUCUGCGUAUUGUUCAUUGAC ..(((.(((((......((.....))..................(((((((((.((((.......)))).)))))))))))))))))((((((...))))))............. ( -26.80) >DroSim_CAF1 25926 115 - 1 AACCGGCCAAAACAUUAGCAUACACCCAUAUUUCCACGAAUCAGUGCUAAAUAGUUUCCGAUAUUGAAAAUAUUUAGCAUUUGGCAGGCGUAGCUUCUGCGUAUUGUUCAUUGAC .....((((((........................((......))((((((((.((((.......)))).)))))))).))))))..((((((...))))))............. ( -23.30) >DroEre_CAF1 25761 113 - 1 AACCGCCAAAAGCAUUAGCAUACACU--UAUUUCCACGAACCAGUGCUAAAUAGUUUCCGAAUUUGAAAAUAUUUAGCAUUUGGCAGGCGUAGCUUCUGCGUAUUGUUCAUUGAC ....((((((.((....)).......--................(((((((((.((((.......)))).)))))))))))))))..((((((...))))))............. ( -26.00) >DroYak_CAF1 45977 113 - 1 AACCGGCAAAAGCAUUAGCAUACACC--UAUUUCCACGAACCAGUGCUAAAUAGUUUCCGAUUUUGGAAAUAUUUAGCAUUUCGCAGGCGUAGCUUCUGCGUAUUGUUCAUUGAC ....(((((..(((..(((.(((.((--(.......((((...((((((((((.((((((....)))))))))))))))))))).))).))))))..)))...)))))....... ( -31.90) >consensus AACCGGCCAAAACAUUAGCAUACACC__UAUUUCCACGAAUCAGUGCUAAAUAGUUUCCGAUAUUGAAAAUAUUUAGCAUUUGGCAGGCGUAGCUUCUGCGUAUUGUUCAUUGAC .....................................((((.(((((((((((.((((.......)))).)))))))))))......((((((...))))))...))))...... (-20.80 = -20.24 + -0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:26 2006