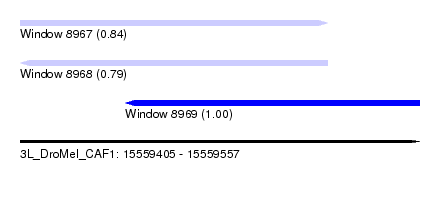
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,559,405 – 15,559,557 |
| Length | 152 |
| Max. P | 0.997002 |

| Location | 15,559,405 – 15,559,522 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 93.50 |
| Mean single sequence MFE | -27.54 |
| Consensus MFE | -23.66 |
| Energy contribution | -23.86 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15559405 117 + 23771897 UUUAUCUUAGUUAUCCUUCUGAUUCUCCUCAGUCUUAGUCUUUGGCCAUACACAGUUAAUAGGCUAUUUGCACAAUUGCCUGGUGGACAAUUGCUGAUGGCCCAGGGAAAACUGUCU .......(((((.(((((((((......))))...........((((((...(((.......((.....)).((((((((....)).))))))))))))))).))))).)))))... ( -30.20) >DroSec_CAF1 21986 117 + 1 UUUGUGUUAUUUAUCCUUCUGAUUCUCCUCCGUCUUAGUCUUUGGCCAUACAUAGUUAAUAGGCUAUUUACACAAUUGCCUGGUGGACAAUUGCUGAUGGCCCAGGGAAAACUGUCU .............(((((((((..(......)..)))).....((((((..((((((....))))))...((((((((((....)).)))))).)))))))).)))))......... ( -25.40) >DroSim_CAF1 22433 117 + 1 UUUGUGUUAUUUAUCCUUCUGAUUCUCCUCCGUCUUAGUCUUUGGCCAUACAUAGUUAAUAGGCUAUUUACACAAUUGCCUGGUGGACAAUUGCUGAUGGCCCAGGGAAAACUGUCU .............(((((((((..(......)..)))).....((((((..((((((....))))))...((((((((((....)).)))))).)))))))).)))))......... ( -25.40) >DroEre_CAF1 22248 117 + 1 UCUAUAUUCCCUAUCCUUAUGAUUCCUCUCCUUCUUAGUCUUUGGCCAUAAAUAGUUAAUAGGCUAUUUACACAAUUGCCUGGUGGACAAUUGCUGAUGGCCCAGGGAAAACUGUCU ......((((((........((((............))))...((((((((((((((....)))))))).((((((((((....)).)))))).)))))))).))))))........ ( -30.50) >DroYak_CAF1 42313 117 + 1 UUUAUAUUCCUUAUCCUUCUGAUUCUUCUGCUUCUUAGUCUUUGGCCAUACAUAGUUAAUAGGCUAUUUACACAAUUGCCUGGUGGACAAUUGCUGAUGGCCCAGGGAAAACUGUCU ......((((((.................(((....)))....((((((..((((((....))))))...((((((((((....)).)))))).)))))))).))))))........ ( -26.20) >consensus UUUAUAUUACUUAUCCUUCUGAUUCUCCUCCGUCUUAGUCUUUGGCCAUACAUAGUUAAUAGGCUAUUUACACAAUUGCCUGGUGGACAAUUGCUGAUGGCCCAGGGAAAACUGUCU ......................................(((((((((((..((((((....))))))...((((((((((....)).)))))).)))))).)))))))......... (-23.66 = -23.86 + 0.20)



| Location | 15,559,405 – 15,559,522 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 93.50 |
| Mean single sequence MFE | -28.26 |
| Consensus MFE | -22.44 |
| Energy contribution | -22.28 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15559405 117 - 23771897 AGACAGUUUUCCCUGGGCCAUCAGCAAUUGUCCACCAGGCAAUUGUGCAAAUAGCCUAUUAACUGUGUAUGGCCAAAGACUAAGACUGAGGAGAAUCAGAAGGAUAACUAAGAUAAA ....((((.(((...(((((((((((((((((.....)))))))))((.....))..........)).))))))...........((((......))))..))).))))........ ( -31.30) >DroSec_CAF1 21986 117 - 1 AGACAGUUUUCCCUGGGCCAUCAGCAAUUGUCCACCAGGCAAUUGUGUAAAUAGCCUAUUAACUAUGUAUGGCCAAAGACUAAGACGGAGGAGAAUCAGAAGGAUAAAUAACACAAA .....(((((((((((((((((.(((((((((.....)))))))))....((((........))))).))))))...........))).)))))))..................... ( -26.70) >DroSim_CAF1 22433 117 - 1 AGACAGUUUUCCCUGGGCCAUCAGCAAUUGUCCACCAGGCAAUUGUGUAAAUAGCCUAUUAACUAUGUAUGGCCAAAGACUAAGACGGAGGAGAAUCAGAAGGAUAAAUAACACAAA .....(((((((((((((((((.(((((((((.....)))))))))....((((........))))).))))))...........))).)))))))..................... ( -26.70) >DroEre_CAF1 22248 117 - 1 AGACAGUUUUCCCUGGGCCAUCAGCAAUUGUCCACCAGGCAAUUGUGUAAAUAGCCUAUUAACUAUUUAUGGCCAAAGACUAAGAAGGAGAGGAAUCAUAAGGAUAGGGAAUAUAGA .....((.(((((((((((....(((((((((.....)))))))))((((((((........))))))))))))...............((....)).......))))))).))... ( -32.40) >DroYak_CAF1 42313 117 - 1 AGACAGUUUUCCCUGGGCCAUCAGCAAUUGUCCACCAGGCAAUUGUGUAAAUAGCCUAUUAACUAUGUAUGGCCAAAGACUAAGAAGCAGAAGAAUCAGAAGGAUAAGGAAUAUAAA .....((.((((((((((((((.(((((((((.....)))))))))....((((........))))).)))))).....((....)))))....(((.....)))..)))).))... ( -24.20) >consensus AGACAGUUUUCCCUGGGCCAUCAGCAAUUGUCCACCAGGCAAUUGUGUAAAUAGCCUAUUAACUAUGUAUGGCCAAAGACUAAGACGGAGGAGAAUCAGAAGGAUAAAUAACAUAAA ....((((((.....((((((..(((((((((.....)))))))))....((((........))))..)))))).)))))).................................... (-22.44 = -22.28 + -0.16)



| Location | 15,559,445 – 15,559,557 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 96.29 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -26.37 |
| Energy contribution | -26.21 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.997002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15559445 112 - 23771897 CC---UCGCUCUUUUCUGCACUUAUCAGUAUUCGCAGCAGACAGUUUUCCCUGGGCCAUCAGCAAUUGUCCACCAGGCAAUUGUGCAAAUAGCCUAUUAACUGUGUAUGGCCAAA ..---.........(((((.................)))))(((......)))(((((((((((((((((.....)))))))))((.....))..........)).))))))... ( -27.33) >DroSec_CAF1 22026 112 - 1 GC---UCGCUCCUUUCUGCACUUAUCAGUAUUCGCAGCAGACAGUUUUCCCUGGGCCAUCAGCAAUUGUCCACCAGGCAAUUGUGUAAAUAGCCUAUUAACUAUGUAUGGCCAAA ((---(.((......(((.......))).....)))))...(((......)))(((((((.(((((((((.....)))))))))....((((........))))).))))))... ( -27.50) >DroSim_CAF1 22473 112 - 1 GC---UCGCUCUUUUCUGCACUUAUCAGUAUUCGCAGCAGACAGUUUUCCCUGGGCCAUCAGCAAUUGUCCACCAGGCAAUUGUGUAAAUAGCCUAUUAACUAUGUAUGGCCAAA ((---(.((......(((.......))).....)))))...(((......)))(((((((.(((((((((.....)))))))))....((((........))))).))))))... ( -27.50) >DroEre_CAF1 22288 112 - 1 CC---UCGCUCAUUUCUGCACUUAUCAGUAUUCGCAGCAGACAGUUUUCCCUGGGCCAUCAGCAAUUGUCCACCAGGCAAUUGUGUAAAUAGCCUAUUAACUAUUUAUGGCCAAA ..---.........(((((.................)))))(((......)))((((....(((((((((.....)))))))))((((((((........))))))))))))... ( -29.43) >DroYak_CAF1 42353 115 - 1 CCUUCUCUCUCAUUUCUGCACUUAUCAGUAUUCGCAGCAGACAGUUUUCCCUGGGCCAUCAGCAAUUGUCCACCAGGCAAUUGUGUAAAUAGCCUAUUAACUAUGUAUGGCCAAA ..............(((((.................)))))(((......)))(((((((.(((((((((.....)))))))))....((((........))))).))))))... ( -26.83) >consensus CC___UCGCUCAUUUCUGCACUUAUCAGUAUUCGCAGCAGACAGUUUUCCCUGGGCCAUCAGCAAUUGUCCACCAGGCAAUUGUGUAAAUAGCCUAUUAACUAUGUAUGGCCAAA ..............(((((.................)))))(((......)))((((((..(((((((((.....)))))))))....((((........))))..))))))... (-26.37 = -26.21 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:14 2006