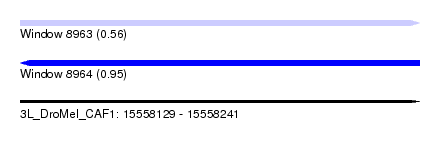
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,558,129 – 15,558,241 |
| Length | 112 |
| Max. P | 0.954011 |

| Location | 15,558,129 – 15,558,241 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.71 |
| Mean single sequence MFE | -30.57 |
| Consensus MFE | -24.98 |
| Energy contribution | -27.03 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
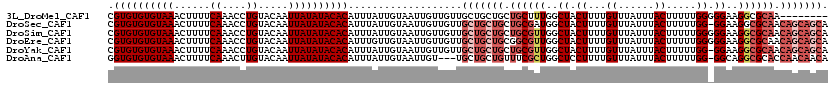
>3L_DroMel_CAF1 15558129 112 + 23771897 CGUGUGUGUAAACUUUUCAAACCUGUACAAUUAUAUACACAUUUAUUGUAAUUGUUGUUGCUGCUGCUGCUUUGGCUACUUUUGUUUAUUUACUUUUUGGGGGAAGGCGCAA-------- .((((((((((......((....)).....)))))))))).......(((((....)))))...(((.(((((..((......((......))......))..)))))))).-------- ( -20.10) >DroSec_CAF1 20725 119 + 1 CGUGUGUGUAAACUUUUCAAACCUGUACAAUUAUAUACACAUUUAUUGUAAUUGUUGUUGCUGCUGCUGCGAUGGCUACUUUUGUUUAUUUACUUUUUGG-GGAAGGCGCAACAGCAGCA .((((((((((......((....)).....))))))))))..................((((((((.((((....(..(....((......)).....).-.)....)))).)))))))) ( -30.80) >DroSim_CAF1 21172 120 + 1 CGUGUGUGUAAACUUUUCAAACCUGUACAAUUAUAUACACAUUUAUUGUAAUUGUUGUUGCUGCUGCUGCGUUGGCUACUUUUGUUUAUUUACUUUUUGGGGGAAGGCGCAACAGCAGCA .((((((((((......((....)).....))))))))))..................((((((((.((((((..(..((...((......)).....))..)..)))))).)))))))) ( -35.20) >DroEre_CAF1 20842 120 + 1 CGUGUGUGUAAACUUUUCAAACCUGUACAAUUAUAUACACAUUUGUUGUAAUUGUUGUUGCUGCUGCGGCGUUGGCUACUUUUGUUUAUUUACUUUUUGGGGGAAGGCGCAACAGCAGCA .((((((((((......((....)).....))))))))))..................((((((((..(((((..(..((...((......)).....))..)..)))))..)))))))) ( -34.90) >DroYak_CAF1 40891 119 + 1 CGUGUGUGUAAACUUUUCAAACCUGUACAAUUAUAUACACAUUUAUUGUAAUUGUUGUUGCUGCUGCUGCGUUGGCUACUUUUGUUUAUUUACUUUUUGG-GGAAGGCGCAACAGCAGCA .((((((((((......((....)).....))))))))))..................((((((((.((((((..(..(....((......)).....).-.)..)))))).)))))))) ( -34.20) >DroAna_CAF1 22419 116 + 1 GGUGUGUGUAAACUUUUCAAACUUGUACAAUUAUAUACACAUUUAUUGUAAUUGU---UGCUGCUGUUUCGCUGGCUCCUUUUGUUUAUUUACUUUUUGG-GGCAGGCGCACCAACAACA .((((((((((......((....)).....))))))))))...........((((---((.(((......(((.(((((....((......)).....))-))).)))))).)))))).. ( -28.20) >consensus CGUGUGUGUAAACUUUUCAAACCUGUACAAUUAUAUACACAUUUAUUGUAAUUGUUGUUGCUGCUGCUGCGUUGGCUACUUUUGUUUAUUUACUUUUUGG_GGAAGGCGCAACAGCAGCA .((((((((((......((....)).....))))))))))...................(((((((.((((((..((.((...((......)).....)).))..)))))).))))))). (-24.98 = -27.03 + 2.06)

| Location | 15,558,129 – 15,558,241 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.71 |
| Mean single sequence MFE | -27.05 |
| Consensus MFE | -22.16 |
| Energy contribution | -23.80 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
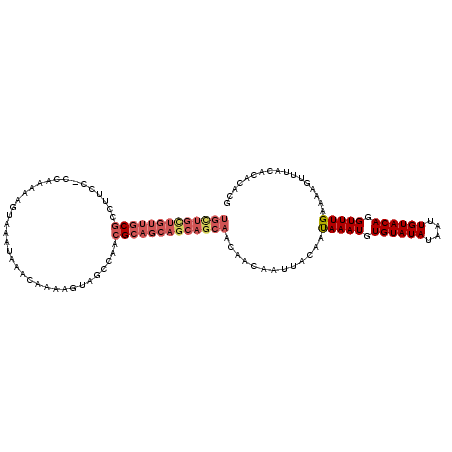
>3L_DroMel_CAF1 15558129 112 - 23771897 --------UUGCGCCUUCCCCCAAAAAGUAAAUAAACAAAAGUAGCCAAAGCAGCAGCAGCAACAACAAUUACAAUAAAUGUGUAUAUAAUUGUACAGGUUUGAAAAGUUUACACACACG --------...................((((((........(((((....)).((....)).........)))..(((((.((((((....)))))).)))))....))))))....... ( -16.40) >DroSec_CAF1 20725 119 - 1 UGCUGCUGUUGCGCCUUCC-CCAAAAAGUAAAUAAACAAAAGUAGCCAUCGCAGCAGCAGCAACAACAAUUACAAUAAAUGUGUAUAUAAUUGUACAGGUUUGAAAAGUUUACACACACG (((((((((((((......-.......((......))............))))))))))))).............(((((.((((((....)))))).)))))................. ( -30.61) >DroSim_CAF1 21172 120 - 1 UGCUGCUGUUGCGCCUUCCCCCAAAAAGUAAAUAAACAAAAGUAGCCAACGCAGCAGCAGCAACAACAAUUACAAUAAAUGUGUAUAUAAUUGUACAGGUUUGAAAAGUUUACACACACG (((((((((((((.(((........))).......((....))......))))))))))))).............(((((.((((((....)))))).)))))................. ( -30.70) >DroEre_CAF1 20842 120 - 1 UGCUGCUGUUGCGCCUUCCCCCAAAAAGUAAAUAAACAAAAGUAGCCAACGCCGCAGCAGCAACAACAAUUACAACAAAUGUGUAUAUAAUUGUACAGGUUUGAAAAGUUUACACACACG (((((((((.(((.(((........))).......((....))......))).))))))))).............(((((.((((((....)))))).)))))................. ( -29.00) >DroYak_CAF1 40891 119 - 1 UGCUGCUGUUGCGCCUUCC-CCAAAAAGUAAAUAAACAAAAGUAGCCAACGCAGCAGCAGCAACAACAAUUACAAUAAAUGUGUAUAUAAUUGUACAGGUUUGAAAAGUUUACACACACG (((((((((((((......-.......((......))............))))))))))))).............(((((.((((((....)))))).)))))................. ( -30.61) >DroAna_CAF1 22419 116 - 1 UGUUGUUGGUGCGCCUGCC-CCAAAAAGUAAAUAAACAAAAGGAGCCAGCGAAACAGCAGCA---ACAAUUACAAUAAAUGUGUAUAUAAUUGUACAAGUUUGAAAAGUUUACACACACC ((((((((...(((..((.-((.....((......))....)).))..)))...))))))))---..........(((((.((((((....)))))).)))))................. ( -25.00) >consensus UGCUGCUGUUGCGCCUUCC_CCAAAAAGUAAAUAAACAAAAGUAGCCAACGCAGCAGCAGCAACAACAAUUACAAUAAAUGUGUAUAUAAUUGUACAGGUUUGAAAAGUUUACACACACG (((((((((((((....................................))))))))))))).............(((((.((((((....)))))).)))))................. (-22.16 = -23.80 + 1.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:09 2006