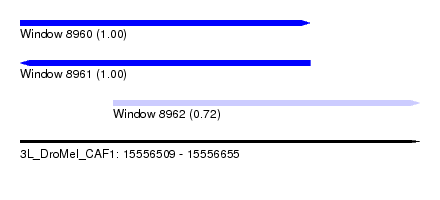
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,556,509 – 15,556,655 |
| Length | 146 |
| Max. P | 0.999341 |

| Location | 15,556,509 – 15,556,615 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -28.66 |
| Consensus MFE | -25.34 |
| Energy contribution | -25.70 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.999341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15556509 106 + 23771897 -----UUGUUCCACGGCACUUGUUGACACAUUUGUGUGGAUUUUCACCAAUUUUCACUUGAGCGUUAAGCGUUCGCUUAACCCUUAGAUGUGCCAGGCAAUUUACAUAAUU -----(((((....(((((..(((.((((....)))).)))................(((((.(((((((....))))))).)))))..))))).)))))........... ( -27.40) >DroSec_CAF1 19182 106 + 1 -----UUGUUCCUCGGCACUUGUUGACACAUUUGUGUGGAUUUUCACCAAUUUUCACUUGAGCGUUAAGCGUUCGCUUAACCCUUAGAUGUGCCAGGCAAUUUACAUAAUU -----..((((((.(((((..(((.((((....)))).)))................(((((.(((((((....))))))).)))))..)))))))).))).......... ( -29.70) >DroSim_CAF1 19669 106 + 1 -----UUGUUCCUCGGCACUUGUUGACACAUUUGUGUGGAUUUUCACCAAUUUUCACUUGAGCGUUAAGCGUUCGCUUAACCCUUAGAUGUGCCAGGCAAUUUACAUAAUU -----..((((((.(((((..(((.((((....)))).)))................(((((.(((((((....))))))).)))))..)))))))).))).......... ( -29.70) >DroEre_CAF1 19286 106 + 1 -----UUGUUUCUCGCCACUUGUUGACACAUUUGUGUGGAUUUUCACCAAUUUUCACUUGAGCGUUAAGCGUUCGCUUAACCCUUAGAUAUGCCAGGCAAUUUGCAUAAUU -----.(((.....(((....(((.((((....)))).)))................(((((.(((((((....))))))).)))))........))).....)))..... ( -24.10) >DroYak_CAF1 39450 111 + 1 GUUUGUUGUUUCUCGGCACUUGUUGACACAUUUGUGUGGAUUUUCACCAAUUUUCACUUGAGCGUUAAGCGUUCGCUUAACCCUUAGAUGUGCCAGGCAAUUUACAUAAUU ((..(((((((...(((((..(((.((((....)))).)))................(((((.(((((((....))))))).)))))..))))))))))))..))...... ( -32.40) >consensus _____UUGUUCCUCGGCACUUGUUGACACAUUUGUGUGGAUUUUCACCAAUUUUCACUUGAGCGUUAAGCGUUCGCUUAACCCUUAGAUGUGCCAGGCAAUUUACAUAAUU ..........(((.(((((..(((.((((....)))).)))................(((((.(((((((....))))))).)))))..)))))))).............. (-25.34 = -25.70 + 0.36)

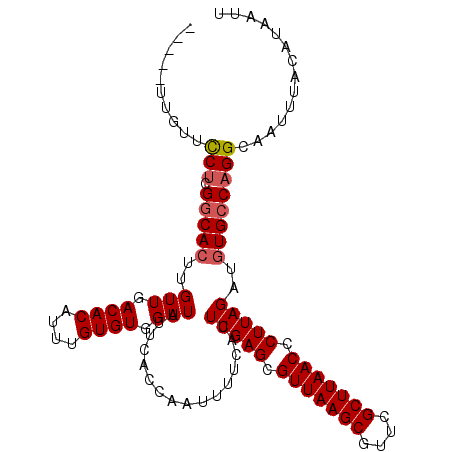
| Location | 15,556,509 – 15,556,615 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -27.84 |
| Consensus MFE | -24.18 |
| Energy contribution | -24.02 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.998737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15556509 106 - 23771897 AAUUAUGUAAAUUGCCUGGCACAUCUAAGGGUUAAGCGAACGCUUAACGCUCAAGUGAAAAUUGGUGAAAAUCCACACAAAUGUGUCAACAAGUGCCGUGGAACAA----- ..............((((((((.....((.(((((((....))))))).))....(((..(((.(((......)))...)))...)))....)))))).)).....----- ( -28.20) >DroSec_CAF1 19182 106 - 1 AAUUAUGUAAAUUGCCUGGCACAUCUAAGGGUUAAGCGAACGCUUAACGCUCAAGUGAAAAUUGGUGAAAAUCCACACAAAUGUGUCAACAAGUGCCGAGGAACAA----- ..............((((((((.....((.(((((((....))))))).))....(((..(((.(((......)))...)))...)))....))))).))).....----- ( -28.90) >DroSim_CAF1 19669 106 - 1 AAUUAUGUAAAUUGCCUGGCACAUCUAAGGGUUAAGCGAACGCUUAACGCUCAAGUGAAAAUUGGUGAAAAUCCACACAAAUGUGUCAACAAGUGCCGAGGAACAA----- ..............((((((((.....((.(((((((....))))))).))....(((..(((.(((......)))...)))...)))....))))).))).....----- ( -28.90) >DroEre_CAF1 19286 106 - 1 AAUUAUGCAAAUUGCCUGGCAUAUCUAAGGGUUAAGCGAACGCUUAACGCUCAAGUGAAAAUUGGUGAAAAUCCACACAAAUGUGUCAACAAGUGGCGAGAAACAA----- .....(((...(((..((((((((...((.(((((((....))))))).))..........((((((......))).))))))))))).)))...)))........----- ( -26.80) >DroYak_CAF1 39450 111 - 1 AAUUAUGUAAAUUGCCUGGCACAUCUAAGGGUUAAGCGAACGCUUAACGCUCAAGUGAAAAUUGGUGAAAAUCCACACAAAUGUGUCAACAAGUGCCGAGAAACAACAAAC .....(((......(.((((((.....((.(((((((....))))))).))....(((..(((.(((......)))...)))...)))....)))))).).....)))... ( -26.40) >consensus AAUUAUGUAAAUUGCCUGGCACAUCUAAGGGUUAAGCGAACGCUUAACGCUCAAGUGAAAAUUGGUGAAAAUCCACACAAAUGUGUCAACAAGUGCCGAGGAACAA_____ ...........(((..((((((((...((.(((((((....))))))).))..........((((((......))).))))))))))).)))................... (-24.18 = -24.02 + -0.16)



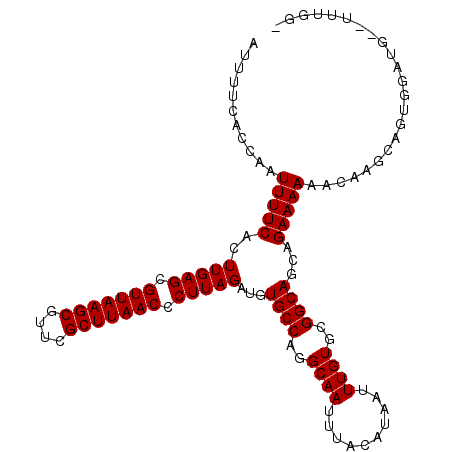
| Location | 15,556,543 – 15,556,655 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 93.46 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -22.82 |
| Energy contribution | -22.82 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15556543 112 + 23771897 AUUUUCACCAAUUUUCACUUGAGCGUUAAGCGUUCGCUUAACCCUUAGAUGUGCCAGGCAAUUUACAUAAUUUGUGCGGCAGCAGAAAAAACAAGCAGUGGAUGUGUUUGGU ......(((((..((((((((((.(((((((....))))))).))))..((((((..((((..........))))..)))).))............)))))).....))))) ( -29.00) >DroSec_CAF1 19216 103 + 1 AUUUUCACCAAUUUUCACUUGAGCGUUAAGCGUUCGCUUAACCCUUAGAUGUGCCAGGCAAUUUACAUAAUUUGUGCGGCAGCAGAAAAAACAAGCAUCGGAU--------- .......((..........((((.(((((((....))))))).))))((((((((..((((..........))))..)))....(......)..)))))))..--------- ( -24.20) >DroSim_CAF1 19703 103 + 1 AUUUUCACCAAUUUUCACUUGAGCGUUAAGCGUUCGCUUAACCCUUAGAUGUGCCAGGCAAUUUACAUAAUUUGUGCGGCAGCAGAAAAAACAAGCAUCGGAU--------- .......((..........((((.(((((((....))))))).))))((((((((..((((..........))))..)))....(......)..)))))))..--------- ( -24.20) >DroEre_CAF1 19320 110 + 1 AUUUUCACCAAUUUUCACUUGAGCGUUAAGCGUUCGCUUAACCCUUAGAUAUGCCAGGCAAUUUGCAUAAUUUGUGCGGCAGCAGAAAAAACAAGCAGUGGAUG--UUUGGU ......(((((..((((((((((.(((((((....))))))).)))))...(((...((.....))....(((.(((....))).)))......))))))))..--.))))) ( -30.70) >DroYak_CAF1 39489 110 + 1 AUUUUCACCAAUUUUCACUUGAGCGUUAAGCGUUCGCUUAACCCUUAGAUGUGCCAGGCAAUUUACAUAAUUUGUGCGGCAGCAGAAAAAACAAGCAGUGGAUG--UUUGGC .......((((..((((((((((.(((((((....))))))).))))..((((((..((((..........))))..)))).))............))))))..--.)))). ( -29.30) >consensus AUUUUCACCAAUUUUCACUUGAGCGUUAAGCGUUCGCUUAACCCUUAGAUGUGCCAGGCAAUUUACAUAAUUUGUGCGGCAGCAGAAAAAACAAGCAGUGGAUG__UUUGG_ ...........(((((..(((((.(((((((....))))))).)))))...((((..((((..........))))..))))...)))))....................... (-22.82 = -22.82 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:07 2006