
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,546,485 – 15,546,587 |
| Length | 102 |
| Max. P | 0.732103 |

| Location | 15,546,485 – 15,546,587 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.81 |
| Mean single sequence MFE | -32.05 |
| Consensus MFE | -18.11 |
| Energy contribution | -18.03 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15546485 102 + 23771897 AGGGAGUUACCAAGAAACUCGGUCGUUGGGAUGCUGUGGUGGCUCAACAGAGAUCUGCGGAGACGCAGGUGAAUGA--AUGCCAAUGCUAACAG------UGACAUACAC ...(((((.......))))).(((((((..(.((..((((..(((....)))(((((((....)))))))......--..))))..)))..)))------))))...... ( -32.50) >DroVir_CAF1 4455 107 + 1 AGGGUGUAAAGAAAAAACUUGGUCGCUGGGACGCAGUCGUGGCCAAGCAGCGUUCUGCCGAGACGCAGGUGAGUAAUGAUUUGAAUAUUAUAAGAAAGUCUGUGCUU--- ..(((....((((....(((((((((...(((...)))))))))))).....)))))))(((.((((((......(((((......))))).......)))))))))--- ( -25.42) >DroSec_CAF1 9294 102 + 1 AGGGAGUCACCAAGAAGCUCGGUCGUUGGGAUGCUGUGGUGGCUCAACAGAGAUCUGCAGAGACGCAGGUUAGUAA--AUGCCAAUGCUAACAG------UGAGAUACAC ...(((((((((...(((...(((.....)))))).))))))))).........((((......))))(((((((.--.......)))))))..------.......... ( -31.10) >DroSim_CAF1 9631 102 + 1 AGGGAGUCACCAAGAAGCUCGGUCGUUGGGAUGCUGUGGUGGCUCAACAGAGAUCUGCAGAGACGCAGGUUAGUAA--AUGCCAAUGCUAACAG------UGAGAUACAC ...(((((((((...(((...(((.....)))))).))))))))).........((((......))))(((((((.--.......)))))))..------.......... ( -31.10) >DroEre_CAF1 9522 98 + 1 AGGGAGUGACCAAGAAGCUCGGUCGCUGGGAUGCAGUGGUGGCUCAACAGAGAUCUGCGGAGACGCAGGUAAGUGU--AUUUCAAUGCUUUCUG------UUAAA----G ...((((.((((.....(((((...)))))......)))).))))((((((.(((((((....)))))))((((((--......))))))))))------))...----. ( -37.40) >DroYak_CAF1 29135 98 + 1 AGGGAGUGACCAAGAAGCUCGGUCGCUGGGAUGCUGUGGUGGCUCAACAGAGAUCUGCGGAGACGCAGGUAAGUGA--AUUUCAAUUCAUACGA------UGAAA----G ...((((.((((((...(((((...)))))...)).)))).))))..((..((((((((....)))))))..((((--((....)))))).)..------))...----. ( -34.80) >consensus AGGGAGUCACCAAGAAGCUCGGUCGCUGGGAUGCUGUGGUGGCUCAACAGAGAUCUGCGGAGACGCAGGUAAGUAA__AUGCCAAUGCUAACAG______UGAGAUA__C .((((((.((((.....(((((...)))))......)))).)))).......((((((......))))))...........))........................... (-18.11 = -18.03 + -0.08)

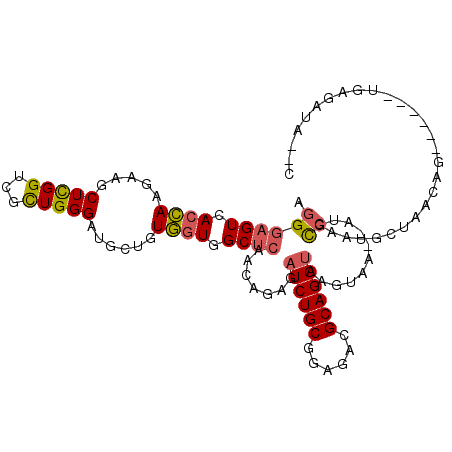
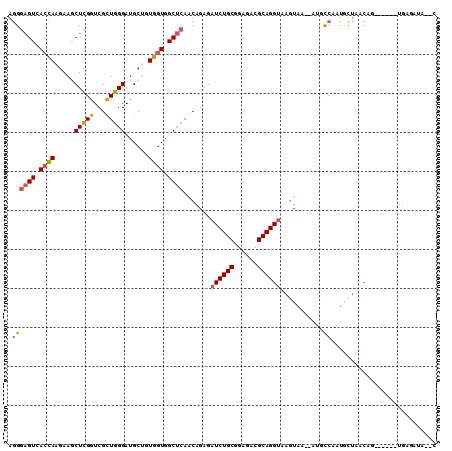
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:00 2006