
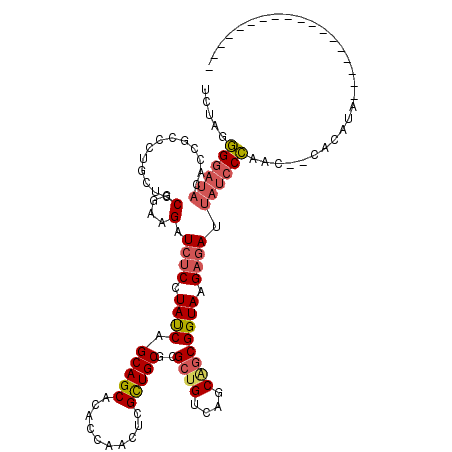
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,534,153 – 15,534,284 |
| Length | 131 |
| Max. P | 0.986379 |

| Location | 15,534,153 – 15,534,253 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.75 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -25.82 |
| Energy contribution | -26.43 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15534153 100 + 23771897 UCUAGGGGAUACACCGCCCUGCUCCGGAAGAUCUCCUAUCAGCAGCACACCAACUCGCUGCGCGCUGUCAGCGGCGGUAAGAGAUUAUGCCAUC--CACAUA------------------ ..((((((......).)))))....((..((((((.((((.(((((..........)))))..((((....)))))))).))))))...))...--......------------------ ( -32.00) >DroSec_CAF1 4203 100 + 1 UCUAGGGGAUACACCGCCCUGCUCCGGAAGAUCUCCUAUCAGCAGCACACCAACUCGCUGCGCGCUGUCAGCGGCGGUAAGAGAUUAUCCCAAC--CACAAA------------------ ..((((((......).)))))....(((.((((((.((((.(((((..........)))))..((((....)))))))).)))))).)))....--......------------------ ( -33.70) >DroAna_CAF1 8434 99 + 1 UCCCGCGGCUACACAGCCCUGCUUCGAAAGAUCUCCUACCAGCAGCACACCAACUCGCUGCGAGGAGUUAGCAGCGGUAAGCCA---CCCGAGCAGCACAUG------------------ ..((((((((....)))).((((((....)).(((((....(((((..........))))).)))))..))))))))...(((.---...).))........------------------ ( -32.90) >DroSim_CAF1 5386 100 + 1 UCUAGGGGAUACACCGCCCUGCUCCGGAAGAUCUCCUAUCAGCAGCACACCAACUCGCUGCGCGCUGUCAGCGGCGGUAAGAGAUUAUCCCAAC--CACAUA------------------ ..((((((......).)))))....(((.((((((.((((.(((((..........)))))..((((....)))))))).)))))).)))....--......------------------ ( -33.70) >DroEre_CAF1 7479 97 + 1 UCUAGGGGAUACACCGCCUUGCUCCGGAAGAUCUCCUAUCAGCAGCACACCAACUCGUUGCGCGCUGUUAGCAGCGGUAAGAGAAUAUCCCCGC--CAC--------------------- ....(((((((..(((........)))....((((.((((.(((((..........)))))..((((....)))))))).)))).)))))))..--...--------------------- ( -34.40) >DroYak_CAF1 5798 118 + 1 UCUAGGGGAUACACCGCCUUGCUCCGGAAGAUCUCCUAUCAGCAGCACACCAACUCGCUGCGCGCUGUCAGCAGCGGUAAGAGAAUAUCCCCAC--CACACAACCAAUUUCUGCUGCUUU ....(((((((..(((........)))....((((.((((.(((((..........)))))..((((....)))))))).)))).)))))))..--........................ ( -36.80) >consensus UCUAGGGGAUACACCGCCCUGCUCCGGAAGAUCUCCUAUCAGCAGCACACCAACUCGCUGCGCGCUGUCAGCAGCGGUAAGAGAUUAUCCCAAC__CACAUA__________________ .....((((((.............(....).((((.((((.(((((..........)))))..((((....)))))))).)))).))))))............................. (-25.82 = -26.43 + 0.61)

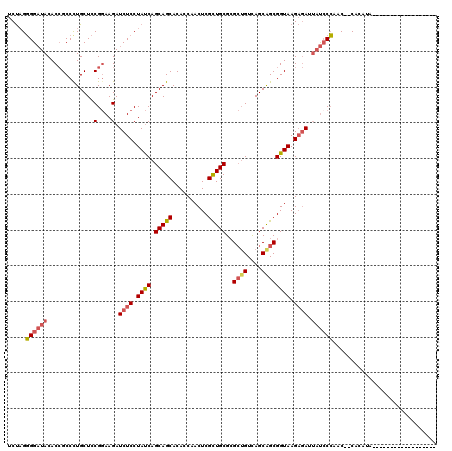
| Location | 15,534,193 – 15,534,284 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 68.19 |
| Mean single sequence MFE | -18.28 |
| Consensus MFE | -15.50 |
| Energy contribution | -15.10 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.54 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15534193 91 + 23771897 AGCAGCACACCAACUCGCUGCGCGCUGUCAGCGGCGGUAAGAGAUUAUGCCAUCCACAUA---------------------------AAAAAACCUCAUGUUGUUUAUGAAAUGACUU (((((((......((((((((.(((.....))))))))..))).(((((.......))))---------------------------)..........)))))))............. ( -20.50) >DroSec_CAF1 4243 73 + 1 AGCAGCACACCAACUCGCUGCGCGCUGUCAGCGGCGGUAAGAGAUUAUCCCAACCACAAA----------------------------AAAA-----------------AAUAGAUUU .(((((..........)))))..((((....))))(((..((.....))...))).....----------------------------....-----------------......... ( -15.80) >DroSim_CAF1 5426 74 + 1 AGCAGCACACCAACUCGCUGCGCGCUGUCAGCGGCGGUAAGAGAUUAUCCCAACCACAUA---------------------------AAAAA-----------------GAAAGAUUU .(((((..........)))))..((((....))))(((..((.....))...))).....---------------------------.....-----------------......... ( -15.80) >DroEre_CAF1 7519 97 + 1 AGCAGCACACCAACUCGUUGCGCGCUGUUAGCAGCGGUAAGAGAAUAUCCCCGCCAC---------------------AAUUCGUUCGAAGAACCUUAUAUUUAUUAUGAAAAGAUUU .(((((..........))))).(((((....)))))(((((.((((...........---------------------.))))((((...)))))))))................... ( -17.70) >DroYak_CAF1 5838 118 + 1 AGCAGCACACCAACUCGCUGCGCGCUGUCAGCAGCGGUAAGAGAAUAUCCCCACCACACAACCAAUUUCUGCUGCUUUAAUUUGUUAGAAGAACCAUUUAUUUGUUAUGAAAAGAUUU (((((((.........(((((.........)))))(((..((.....))...)))..............)))))))....................(((((.....)))))....... ( -21.60) >consensus AGCAGCACACCAACUCGCUGCGCGCUGUCAGCGGCGGUAAGAGAUUAUCCCAACCACAUA___________________________AAAAAACC___U_UU__UUAUGAAAAGAUUU .(((((..........))))).(((((....))))).................................................................................. (-15.50 = -15.10 + -0.40)


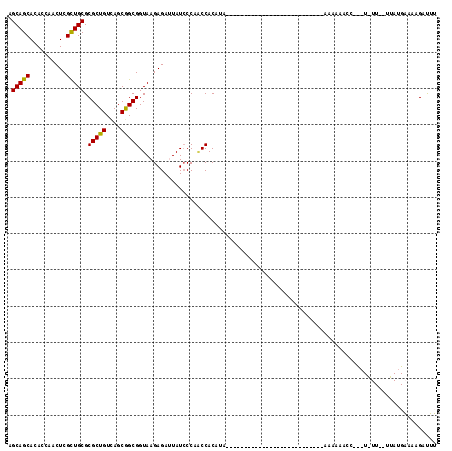
| Location | 15,534,193 – 15,534,284 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 68.19 |
| Mean single sequence MFE | -24.28 |
| Consensus MFE | -17.16 |
| Energy contribution | -16.84 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.18 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15534193 91 - 23771897 AAGUCAUUUCAUAAACAACAUGAGGUUUUUU---------------------------UAUGUGGAUGGCAUAAUCUCUUACCGCCGCUGACAGCGCGCAGCGAGUUGGUGUGCUGCU ..(((((((((((((.(((.....)))..))---------------------------)))).))))))).............(((((..((.((.....))..))..))).)).... ( -23.10) >DroSec_CAF1 4243 73 - 1 AAAUCUAUU-----------------UUUU----------------------------UUUGUGGUUGGGAUAAUCUCUUACCGCCGCUGACAGCGCGCAGCGAGUUGGUGUGCUGCU .........-----------------....----------------------------...(((((.(((.....)))..)))))......((((((((.........)))))))).. ( -19.60) >DroSim_CAF1 5426 74 - 1 AAAUCUUUC-----------------UUUUU---------------------------UAUGUGGUUGGGAUAAUCUCUUACCGCCGCUGACAGCGCGCAGCGAGUUGGUGUGCUGCU .........-----------------.....---------------------------...(((((.(((.....)))..)))))......((((((((.........)))))))).. ( -19.60) >DroEre_CAF1 7519 97 - 1 AAAUCUUUUCAUAAUAAAUAUAAGGUUCUUCGAACGAAUU---------------------GUGGCGGGGAUAUUCUCUUACCGCUGCUAACAGCGCGCAACGAGUUGGUGUGCUGCU ........................((((...)))).....---------------------(..(((((((.....)))..))))..)...((((((((((....)).)))))))).. ( -23.50) >DroYak_CAF1 5838 118 - 1 AAAUCUUUUCAUAACAAAUAAAUGGUUCUUCUAACAAAUUAAAGCAGCAGAAAUUGGUUGUGUGGUGGGGAUAUUCUCUUACCGCUGCUGACAGCGCGCAGCGAGUUGGUGUGCUGCU ..........................................(((((((..((((.((((((((((((((......)))))))((((....))))))))))).))))....))))))) ( -35.60) >consensus AAAUCUUUUCAUAA__AA_A___GGUUUUUC___________________________UAUGUGGUUGGGAUAAUCUCUUACCGCCGCUGACAGCGCGCAGCGAGUUGGUGUGCUGCU .............................................................(((((.((............)))))))...((((((((((....)).)))))))).. (-17.16 = -16.84 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:54 2006