
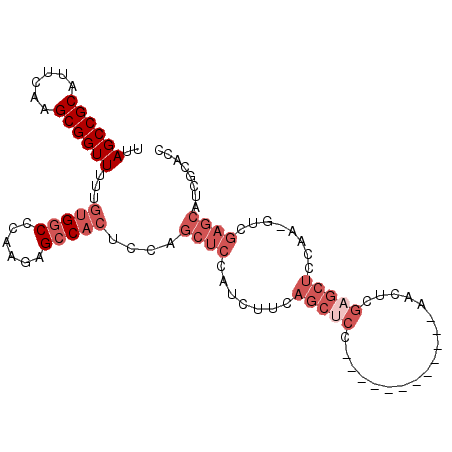
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,509,222 – 15,509,312 |
| Length | 90 |
| Max. P | 0.777006 |

| Location | 15,509,222 – 15,509,312 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.43 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -16.40 |
| Energy contribution | -18.56 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15509222 90 + 23771897 UUAGCCGCAUUCAAGCGGUUUUUGUGGCCCAAGAGCCACUCCAGCUCCAUCUUCAGCGCC------------AACUUGAGCUCCAAACUCGAGCAUCGCACC ..((((((......))))))...(((((......)))))................(((..------------..((((((.......))))))...)))... ( -25.40) >DroSec_CAF1 11984 89 + 1 UUAGCCGCAUUCAAGCGGUUUUUGUGGCCCAAGAGCCACUCCAGCUCCAUCUUCAGCUCC------------AACUUGAGCUCCAA-GUCGAGCAUCGCACC ..((((((......))))))...(((((......)))))....((((...(((.(((((.------------.....)))))..))-)..))))........ ( -28.50) >DroSim_CAF1 7563 89 + 1 UUAGCCGCAUUCAAGCGGUUUUUGUGGCCCAAGAGCCACUCCAGCUCCAUCUUCAGCUCC------------AACUUGAGCUCCAA-GUCGAGCAUCGCACC ..((((((......))))))...(((((......)))))....((((...(((.(((((.------------.....)))))..))-)..))))........ ( -28.50) >DroEre_CAF1 20215 89 + 1 UUAGCCGCAUUCAAGCGGUUUUUGUGGCCCAAGAGCCACUCCAGCUCCAUCUUCAGCUCC------------AACUCGAGCUCCGA-GUCGAGCAUCGCACC ..((((((......))))))...(((((......)))))....((((...(((.(((((.------------.....)))))..))-)..))))........ ( -29.00) >DroYak_CAF1 20267 101 + 1 UUAGCCGCAUUCAAGCGGUUUUUGUGGCCCAAGAGCCACUCCAGCUCCAUCUUCAGCUCCAACUUGAGCUCCAACUCGAGCUCCAA-GUCGAGCAUCGCACC ..((((((......))))))...(((((......)))))................((((..((((((((((......))))).)))-)).))))........ ( -34.10) >DroAna_CAF1 21029 68 + 1 UUAGCCGC--UCAAGCGGUUUUUGCGGCUUAAGAGGCA-----------ACUUCAGC----------------ACUCGAAGCC-----UGAAGCACCGCACC ........--....((((((((((.(((((..(((((.-----------......))----------------.))).)))))-----))))).)))))... ( -21.70) >consensus UUAGCCGCAUUCAAGCGGUUUUUGUGGCCCAAGAGCCACUCCAGCUCCAUCUUCAGCUCC____________AACUCGAGCUCCAA_GUCGAGCAUCGCACC ..((((((......))))))...(((((......)))))....((((.......(((((..................)))))........))))........ (-16.40 = -18.56 + 2.17)


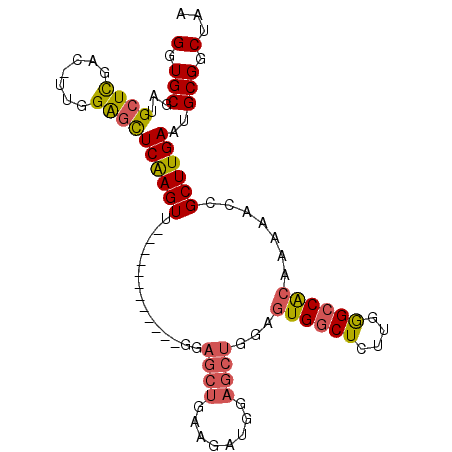
| Location | 15,509,222 – 15,509,312 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.43 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -22.37 |
| Energy contribution | -22.63 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15509222 90 - 23771897 GGUGCGAUGCUCGAGUUUGGAGCUCAAGUU------------GGCGCUGAAGAUGGAGCUGGAGUGGCUCUUGGGCCACAAAAACCGCUUGAAUGCGGCUAA (((((.(.(((.((((.....)))).))))------------.))))).....(((.......((((((....)))))).....((((......))))))). ( -31.00) >DroSec_CAF1 11984 89 - 1 GGUGCGAUGCUCGAC-UUGGAGCUCAAGUU------------GGAGCUGAAGAUGGAGCUGGAGUGGCUCUUGGGCCACAAAAACCGCUUGAAUGCGGCUAA (.((((..((((...-...))))((((((.------------((((((........))))...((((((....)))))).....)))))))).)))).)... ( -32.40) >DroSim_CAF1 7563 89 - 1 GGUGCGAUGCUCGAC-UUGGAGCUCAAGUU------------GGAGCUGAAGAUGGAGCUGGAGUGGCUCUUGGGCCACAAAAACCGCUUGAAUGCGGCUAA (.((((..((((...-...))))((((((.------------((((((........))))...((((((....)))))).....)))))))).)))).)... ( -32.40) >DroEre_CAF1 20215 89 - 1 GGUGCGAUGCUCGAC-UCGGAGCUCGAGUU------------GGAGCUGAAGAUGGAGCUGGAGUGGCUCUUGGGCCACAAAAACCGCUUGAAUGCGGCUAA (((.(((.(((((((-(((.....))))))------------.))))........(((((.....)))))))).))).......((((......)))).... ( -32.40) >DroYak_CAF1 20267 101 - 1 GGUGCGAUGCUCGAC-UUGGAGCUCGAGUUGGAGCUCAAGUUGGAGCUGAAGAUGGAGCUGGAGUGGCUCUUGGGCCACAAAAACCGCUUGAAUGCGGCUAA (((.(((.(((((((-(((.(((((......))))))))))).))))........(((((.....)))))))).))).......((((......)))).... ( -39.80) >DroAna_CAF1 21029 68 - 1 GGUGCGGUGCUUCA-----GGCUUCGAGU----------------GCUGAAGU-----------UGCCUCUUAAGCCGCAAAAACCGCUUGA--GCGGCUAA ..((((((((((((-----(((.....))----------------.)))))))-----------..........))))))....((((....--)))).... ( -22.80) >consensus GGUGCGAUGCUCGAC_UUGGAGCUCAAGUU____________GGAGCUGAAGAUGGAGCUGGAGUGGCUCUUGGGCCACAAAAACCGCUUGAAUGCGGCUAA (.(((...((((.......))))((((((...............((((........))))...((((((....)))))).......))))))..))).)... (-22.37 = -22.63 + 0.27)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:45 2006