
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,504,146 – 15,504,252 |
| Length | 106 |
| Max. P | 0.598480 |

| Location | 15,504,146 – 15,504,252 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.76 |
| Mean single sequence MFE | -18.08 |
| Consensus MFE | -5.96 |
| Energy contribution | -6.30 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15504146 106 + 23771897 CUCUUCAAGCA-UAUCAACAAUCUUAUCUCACCGUAUUUUGUAACCAAAAAAAU-AA-AGCAAAAAAAAAAAAAGUAAACUUGCGCCCAGAUAAGGCCUGAAAUGCCCA ........(((-(.(((....((((((((...(((.((((((............-..-.)))))).......(((....))))))...))))))))..))).))))... ( -16.66) >DroPse_CAF1 13986 89 + 1 CUCUUGAGGCA-UAUCAAGAAUCUUAUCACAAAGUAUUUUGUAACCAAA-AAGC-AAAAGCGAAAAAAAA-----------------CAGAUAAGGCCUGAAAUGCUUC .....((((((-(.(((.(...(((((((((((....))))).......-..((-....)).........-----------------..)))))).).))).))))))) ( -18.90) >DroSec_CAF1 2406 105 + 1 CUCUUCAAGCA-UAUCAACAAUCUUAUCACACAGUAUUUUGUAACCAAA-AAAU-AA-AGCGAAAACAAAAAAAGUAAACUUGCGCCCAGAUAAGGCCUGAAAUGCCCA ........(((-(.(((....(((((((......((((((........)-))))-).-.(((..........(((....))).)))...)))))))..))).))))... ( -16.00) >DroEre_CAF1 15238 104 + 1 CUCUUCAAGCA-UAUCAACAAUCUUAUCUCGCAGUAUUUUGUAACCAAA-AAAU-AA-AGCAAAAA-AAUAAAAGUAAACUUGCGCCGAGAUAAGGCCUGAAAUGCCCA ........(((-(.(((....(((((((((((..((((((........)-))))-).-.)).....-.......(((....)))...)))))))))..))).))))... ( -22.80) >DroAna_CAF1 14643 106 + 1 CUCUUCAGUCAUUAUCAACAAUCUUAUCUUAAAGUAUUUUGUAACCAAA-AAAACAAAAGGAAAAA-AAUAAG-ACCUGCCUGCGCCCAGAUAAGGUCUGAAAUGCCCA ...(((((.(.(((((.....((((((.........((((((.......-...)))))).......-.)))))-)...((....))...))))).).)))))....... ( -15.23) >DroPer_CAF1 13971 88 + 1 CUCUUGAGGCA-UAUCAAGAAUCUUAUCACAAAGUAUUUUGUAACCAAA-AAGC-AAAAGCGAAA-AAAA-----------------CAGAUAAGGCCUGAAAUGCUUC .....((((((-(.(((.(...(((((((((((....))))).......-..((-....))....-....-----------------..)))))).).))).))))))) ( -18.90) >consensus CUCUUCAAGCA_UAUCAACAAUCUUAUCACAAAGUAUUUUGUAACCAAA_AAAU_AA_AGCAAAAAAAAAAAA_GUAAACUUGCGCCCAGAUAAGGCCUGAAAUGCCCA ........(((...(((....(((((((........((((((.................)))))).........((......)).....)))))))..)))..)))... ( -5.96 = -6.30 + 0.33)

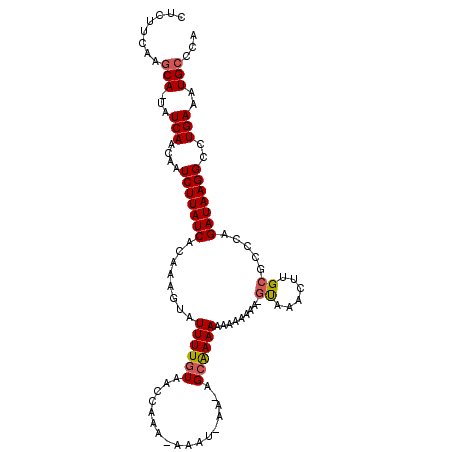
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:42 2006