| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
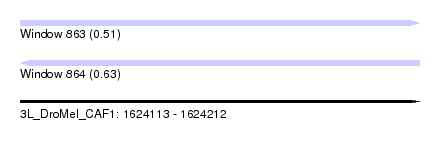
| Location | 1,624,113 – 1,624,212 |
| Length | 99 |
| Max. P | 0.634514 |

| Location | 1,624,113 – 1,624,212 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 81.26 |
| Mean single sequence MFE | -45.68 |
| Consensus MFE | -35.45 |
| Energy contribution | -35.23 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
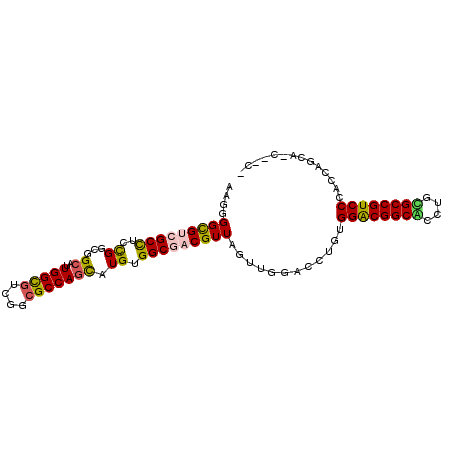
>3L_DroMel_CAF1 1624113 99 + 23771897 ACGGGGCGUGGCCUCCGGAGGCAUUGGCGUUGGCGCCAGCAUGUGGCGUCGUUAGUUGGACCUGUGGACGGCUCCUGGGCCGUCCCAUCCGCAGCUGCC .((((((...))).)))..((((...((...(((((((.....))))))).......(((..((.((((((((....)))))))))))))))...)))) ( -48.10) >DroVir_CAF1 8115 95 + 1 ACGUGGCGUCGCCUCCGGCGGCAUUGGCGUCGGCGCCAGCAUGUGGCGACGUUAGUUGGACUUUUGGACGGCAACGACGCCGUCCCAGCACCAGC---- ...((((((((((..((...((..(((((....))))))).)).))))))))))(((((......(((((((......))))))).....)))))---- ( -45.70) >DroPse_CAF1 8959 97 + 1 CAGGGGCGUCGCCUCCGGCGGCAUUGGUGUGGGCGCCAGCAUGUGGCGACGUUAGUUGGACCUGUGGACGGCGCCAGCGCCGUCCCUCCAACA-CGAC- ....(((((((((..((...((..(((((....))))))).)).))))))))).((((((...(.((((((((....))))))))))))))).-....- ( -49.00) >DroSec_CAF1 8757 99 + 1 AAGAGGCGUGGCCUCCGGAGGCAUUGGCGUCGGCGCCAGCAUGUGGCGUCGUUAGUUGGACCUGUGGGCGGCUCCUGGGCCGUCCCAUCCGCAGCUGCC ..(((((...)))))....((((...((..((((((((.....))))))))......(((..((.((((((((....)))))))))))))))...)))) ( -47.40) >DroWil_CAF1 8405 92 + 1 ACGGGGUGUCGCUUCUGGCGGAAUUGGUGUAGGAGCCAGUAUGUGGCGACGUUAGUUAGACCUUUGGACGGCAACUGUGCCGUCCCAGUCGC------- (((...((((((..(((((...............)))))...)))))).)))......(((....((((((((....))))))))..)))..------- ( -34.86) >DroPer_CAF1 8844 97 + 1 CAGGGGCGUCGCCUCCGGCGGCAUUGGUGUGGGCGCCAGCAUGUGGCGACGUUAGUUGGACCUGUGGACGGCGCCAGCGCCGUCCCUCCAACA-CGAC- ....(((((((((..((...((..(((((....))))))).)).))))))))).((((((...(.((((((((....))))))))))))))).-....- ( -49.00) >consensus AAGGGGCGUCGCCUCCGGCGGCAUUGGCGUCGGCGCCAGCAUGUGGCGACGUUAGUUGGACCUGUGGACGGCACCUGCGCCGUCCCACCAGCA_C__C_ ....(((((((((..((...((..(((((....))))))).)).)))))))))............((((((((....)))))))).............. (-35.45 = -35.23 + -0.22)

| Location | 1,624,113 – 1,624,212 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 81.26 |
| Mean single sequence MFE | -42.11 |
| Consensus MFE | -27.50 |
| Energy contribution | -29.17 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
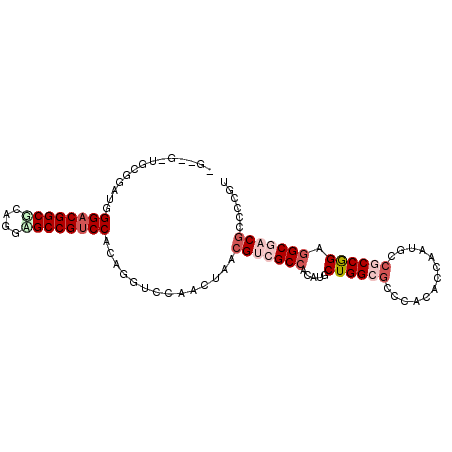
>3L_DroMel_CAF1 1624113 99 - 23771897 GGCAGCUGCGGAUGGGACGGCCCAGGAGCCGUCCACAGGUCCAACUAACGACGCCACAUGCUGGCGCCAACGCCAAUGCCUCCGGAGGCCACGCCCCGU ((((...((((((.(((((((......)))))))....)))).......(.(((((.....))))).)...))...))))..(((.(((...)))))). ( -42.50) >DroVir_CAF1 8115 95 - 1 ----GCUGGUGCUGGGACGGCGUCGUUGCCGUCCAAAAGUCCAACUAACGUCGCCACAUGCUGGCGCCGACGCCAAUGCCGCCGGAGGCGACGCCACGU ----..(((.(((.((((((((....))))))))...)))))).....(((((((.....((((((.((.......)).)))))).)))))))...... ( -42.40) >DroPse_CAF1 8959 97 - 1 -GUCG-UGUUGGAGGGACGGCGCUGGCGCCGUCCACAGGUCCAACUAACGUCGCCACAUGCUGGCGCCCACACCAAUGCCGCCGGAGGCGACGCCCCUG -....-.((((((.((((((((....)))))))).....))))))...(((((((.....((((((..((......)).)))))).)))))))...... ( -45.70) >DroSec_CAF1 8757 99 - 1 GGCAGCUGCGGAUGGGACGGCCCAGGAGCCGCCCACAGGUCCAACUAACGACGCCACAUGCUGGCGCCGACGCCAAUGCCUCCGGAGGCCACGCCUCUU ((((...(((((((((.((((......))))))))....)))......((.(((((.....))))).))..))...))))...((((((...)))))). ( -42.20) >DroWil_CAF1 8405 92 - 1 -------GCGACUGGGACGGCACAGUUGCCGUCCAAAGGUCUAACUAACGUCGCCACAUACUGGCUCCUACACCAAUUCCGCCAGAAGCGACACCCCGU -------..((((.((((((((....))))))))...))))........(((((......(((((...............)))))..)))))....... ( -34.16) >DroPer_CAF1 8844 97 - 1 -GUCG-UGUUGGAGGGACGGCGCUGGCGCCGUCCACAGGUCCAACUAACGUCGCCACAUGCUGGCGCCCACACCAAUGCCGCCGGAGGCGACGCCCCUG -....-.((((((.((((((((....)))))))).....))))))...(((((((.....((((((..((......)).)))))).)))))))...... ( -45.70) >consensus _G__G_UGCGGAUGGGACGGCGCAGGAGCCGUCCACAGGUCCAACUAACGUCGCCACAUGCUGGCGCCCACACCAAUGCCGCCGGAGGCGACGCCCCGU ..............((((((((....))))))))..............(((((((.....((((((.............)))))).)))))))...... (-27.50 = -29.17 + 1.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:27 2006