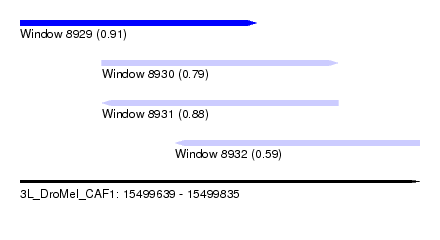
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,499,639 – 15,499,835 |
| Length | 196 |
| Max. P | 0.910790 |

| Location | 15,499,639 – 15,499,755 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.40 |
| Mean single sequence MFE | -40.50 |
| Consensus MFE | -38.61 |
| Energy contribution | -38.17 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15499639 116 + 23771897 GCGAGUGUUACCAAGCGGCAGCAGCCAAAGUCGGGUCUGGUUGCAGAGCGGGGUACACCCCCCG---U-CCUAUUCCAUCGUGCAUUCGGGCCAAAUCAAAUACUCGCAGCAACAACACA (((((((((.......(((....))).......((((((..((((((((((((......)))))---)-.........)).))))..))))))......)))))))))............ ( -43.30) >DroSec_CAF1 40780 117 + 1 GUGAGUGUUACCAAGCGGCAGCAGCCAAAGUCGGGUCUGGUUGCAGAGCGGGGAAGACCCCCCA---UACCUACACCAUCGUGCAUUCGGGCCAAAUCAAAUACUCGCAGCAACAACACA (((((((((.......(((....))).......((((((..(((.....((((......)))).---.....((......)))))..))))))......)))))))))............ ( -37.80) >DroSim_CAF1 41032 120 + 1 GCGAGUGUUACCAAGCGGCAGCAGCCAAAGUCGGGUCUGGUUGCAGAGCGGGGAACACCCCCCACCCUACCUACACCAUCGUGCAUUCGGGCCAAAUCAAAUACUCGCAGCAACAACACA (((((((((.......(((....))).......((((((..(((((.(.((((......)))).).))....((......)))))..))))))......)))))))))............ ( -40.40) >consensus GCGAGUGUUACCAAGCGGCAGCAGCCAAAGUCGGGUCUGGUUGCAGAGCGGGGAACACCCCCCA___UACCUACACCAUCGUGCAUUCGGGCCAAAUCAAAUACUCGCAGCAACAACACA (((((((((.......(((....))).......((((((..(((.....((((......)))).........((......)))))..))))))......)))))))))............ (-38.61 = -38.17 + -0.44)


| Location | 15,499,679 – 15,499,795 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.96 |
| Mean single sequence MFE | -24.77 |
| Consensus MFE | -22.53 |
| Energy contribution | -22.53 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15499679 116 + 23771897 UUGCAGAGCGGGGUACACCCCCCG---U-CCUAUUCCAUCGUGCAUUCGGGCCAAAUCAAAUACUCGCAGCAACAACACAAAAUCUAUAAUAAAAAACAGGUUAACAAAUGUUGCCGCAA ..((((..(((((......)))))---.-.))......(((......)))))..............((.((((((......(((((............)))))......)))))).)).. ( -25.50) >DroSec_CAF1 40820 117 + 1 UUGCAGAGCGGGGAAGACCCCCCA---UACCUACACCAUCGUGCAUUCGGGCCAAAUCAAAUACUCGCAGCAACAACACAAAAUCUAUAAUAAAAAACAGGUUAACAAAUGUUGCCGCAA .....(((.((((......)))).---..(((.(((....))).....)))............)))((.((((((......(((((............)))))......)))))).)).. ( -23.00) >DroSim_CAF1 41072 120 + 1 UUGCAGAGCGGGGAACACCCCCCACCCUACCUACACCAUCGUGCAUUCGGGCCAAAUCAAAUACUCGCAGCAACAACACAAAAUCUAUAAUAAAAAACAGGUUAACAAAUGUUGCCGCAA .....(((.((((......)))).(((......(((....))).....)))............)))((.((((((......(((((............)))))......)))))).)).. ( -25.80) >consensus UUGCAGAGCGGGGAACACCCCCCA___UACCUACACCAUCGUGCAUUCGGGCCAAAUCAAAUACUCGCAGCAACAACACAAAAUCUAUAAUAAAAAACAGGUUAACAAAUGUUGCCGCAA .....(((.((((......)))).................(.((......)))..........)))((.((((((......(((((............)))))......)))))).)).. (-22.53 = -22.53 + 0.00)

| Location | 15,499,679 – 15,499,795 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.96 |
| Mean single sequence MFE | -39.50 |
| Consensus MFE | -33.92 |
| Energy contribution | -34.37 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15499679 116 - 23771897 UUGCGGCAACAUUUGUUAACCUGUUUUUUAUUAUAGAUUUUGUGUUGUUGCUGCGAGUAUUUGAUUUGGCCCGAAUGCACGAUGGAAUAGG-A---CGGGGGGUGUACCCCGCUCUGCAA (((((((((((......((.((((........)))).))......)))))))))))(((((((........))))))).........(((.-.---(((((......)))))..)))... ( -37.60) >DroSec_CAF1 40820 117 - 1 UUGCGGCAACAUUUGUUAACCUGUUUUUUAUUAUAGAUUUUGUGUUGUUGCUGCGAGUAUUUGAUUUGGCCCGAAUGCACGAUGGUGUAGGUA---UGGGGGGUCUUCCCCGCUCUGCAA (((((((((((......((.((((........)))).))......)))))))))))(((((((........))))))).......((((((..---.(.((((....)))).))))))). ( -36.90) >DroSim_CAF1 41072 120 - 1 UUGCGGCAACAUUUGUUAACCUGUUUUUUAUUAUAGAUUUUGUGUUGUUGCUGCGAGUAUUUGAUUUGGCCCGAAUGCACGAUGGUGUAGGUAGGGUGGGGGGUGUUCCCCGCUCUGCAA (((((((((((......((.((((........)))).))......)))))))))))(((((((........)))))))............((((((((((((....)))))))))))).. ( -44.00) >consensus UUGCGGCAACAUUUGUUAACCUGUUUUUUAUUAUAGAUUUUGUGUUGUUGCUGCGAGUAUUUGAUUUGGCCCGAAUGCACGAUGGUGUAGGUA___UGGGGGGUGUUCCCCGCUCUGCAA (((((((((((......((.((((........)))).))......)))))))))))(((((((........))))))).......((((((.....(((((......))))).)))))). (-33.92 = -34.37 + 0.45)


| Location | 15,499,715 – 15,499,835 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -34.70 |
| Energy contribution | -34.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15499715 120 - 23771897 CAAGUUUCUUCCGACGCUAUGGGCUGCGUCGACGUCAGCGUUGCGGCAACAUUUGUUAACCUGUUUUUUAUUAUAGAUUUUGUGUUGUUGCUGCGAGUAUUUGAUUUGGCCCGAAUGCAC ...(((......)))((..(((((((.(((((((....))))(((((((((......((.((((........)))).))......)))))))))........))).)))))))...)).. ( -34.70) >DroSec_CAF1 40857 120 - 1 CAAGUUUCUUCCGACGCUAUGGGCUGCGUCGACGUCAGCGUUGCGGCAACAUUUGUUAACCUGUUUUUUAUUAUAGAUUUUGUGUUGUUGCUGCGAGUAUUUGAUUUGGCCCGAAUGCAC ...(((......)))((..(((((((.(((((((....))))(((((((((......((.((((........)))).))......)))))))))........))).)))))))...)).. ( -34.70) >DroSim_CAF1 41112 120 - 1 CAAGUUUCUUCCGACGCUAUGGGCUGCGUCGACGUCAGCGUUGCGGCAACAUUUGUUAACCUGUUUUUUAUUAUAGAUUUUGUGUUGUUGCUGCGAGUAUUUGAUUUGGCCCGAAUGCAC ...(((......)))((..(((((((.(((((((....))))(((((((((......((.((((........)))).))......)))))))))........))).)))))))...)).. ( -34.70) >consensus CAAGUUUCUUCCGACGCUAUGGGCUGCGUCGACGUCAGCGUUGCGGCAACAUUUGUUAACCUGUUUUUUAUUAUAGAUUUUGUGUUGUUGCUGCGAGUAUUUGAUUUGGCCCGAAUGCAC ...(((......)))((..(((((((.(((((((....))))(((((((((......((.((((........)))).))......)))))))))........))).)))))))...)).. (-34.70 = -34.70 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:38 2006