
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,496,976 – 15,497,215 |
| Length | 239 |
| Max. P | 0.935379 |

| Location | 15,496,976 – 15,497,096 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.10 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -32.39 |
| Energy contribution | -32.50 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.55 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15496976 120 + 23771897 UAUAAUUAGCGAUAAGAAAAUGUUGUGACCCGCUGGGAAAGAAAGGCACAGCAUAAGAGUCGAUAUGGAGUGACUCAUUUUACCAACGCGAUAAGCGGUCCUUUGUGUCCUGCAAGGGAA ..........(.(((((..((((((((.((..((.....))...))))))))))..((((((........)))))).))))).)..(((.....))).((((((((.....)))))))). ( -34.60) >DroSec_CAF1 38160 119 + 1 AAUAAUUAGCGAUAAGAAAAUGUUGUGACCCGCUGGGAAAGA-AGGCACAGCAUAAGAGUCGAUACGGAAUGACUCAUUUUACCAACGCGAUAAGCGGUCCUUUGUGUCAUGCAGGAGAA ..........(.(((((..((((((((.((..((.....)).-.))))))))))..((((((........)))))).))))).)..(((.....))).((..((((.....))))..)). ( -27.80) >DroSim_CAF1 38356 119 + 1 AAUAAUUAGCGAUAAGAAAAUGUUGUGACCCGCUGGGAAAGA-AGGCACAGCAUAAGAGUCGAUAUGGAGUGACUCAUUUUACCAACGCGAUAAGCGGUCCUUUGUGUCCUGCAAGGGAA ..........(.(((((..((((((((.((..((.....)).-.))))))))))..((((((........)))))).))))).)..(((.....))).((((((((.....)))))))). ( -35.10) >consensus AAUAAUUAGCGAUAAGAAAAUGUUGUGACCCGCUGGGAAAGA_AGGCACAGCAUAAGAGUCGAUAUGGAGUGACUCAUUUUACCAACGCGAUAAGCGGUCCUUUGUGUCCUGCAAGGGAA ..........(.(((((..((((((((.((..((.....))...))))))))))..((((((........)))))).))))).)..(((.....))).((((((((.....)))))))). (-32.39 = -32.50 + 0.11)



| Location | 15,497,056 – 15,497,176 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.22 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -23.72 |
| Energy contribution | -24.17 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15497056 120 + 23771897 UACCAACGCGAUAAGCGGUCCUUUGUGUCCUGCAAGGGAAAUCCUUAUCAGCAUACACAUAUGUAUGUUUAGUGUGGAACUAGCAUUCCUCUUAAAGAUUAUUAGACAGAGCCUUUAAAA ..(((.(((((((((...((((((((.....))))))))....))))))((((((((....))))))))..))))))..............((((((.((........))..)))))).. ( -30.70) >DroSec_CAF1 38239 104 + 1 UACCAACGCGAUAAGCGGUCCUUUGUGUCAUGCAGGAGAAAUCCUUAUCAGCAAACACAUAUGUAUGUUUAGUGAGGAACUAGCAUUCCUCUUAAAGAUUAUUA---------------- ......(((.....)))......(((((..(((((((....)))).....))).))))).....((.(((((.((((((......))))))))))).)).....---------------- ( -24.90) >DroSim_CAF1 38435 120 + 1 UACCAACGCGAUAAGCGGUCCUUUGUGUCCUGCAAGGGAAAUCCUUAUCAGCACACACAUAUGUAUGUUUAGUGAGGAACUAGCAUUCCUCUUAAAGAUUAUUACACAGAGGCUUUAAAC ......(((.....)))(.(((((((((.(((..((((....))))..))).............((.(((((.((((((......))))))))))).))....))))))))))....... ( -30.10) >consensus UACCAACGCGAUAAGCGGUCCUUUGUGUCCUGCAAGGGAAAUCCUUAUCAGCAAACACAUAUGUAUGUUUAGUGAGGAACUAGCAUUCCUCUUAAAGAUUAUUA_ACAGAG_CUUUAAA_ .......((((((((...((((((((.....))))))))....)))))).))............((.(((((.((((((......))))))))))).))..................... (-23.72 = -24.17 + 0.45)



| Location | 15,497,096 – 15,497,215 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.39 |
| Mean single sequence MFE | -25.37 |
| Consensus MFE | -21.45 |
| Energy contribution | -21.57 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15497096 119 - 23771897 ACCACGGCUUAGAGCCGUAC-AAUAUUUUAAUUUCGAGGAUUUUAAAGGCUCUGUCUAAUAAUCUUUAAGAGGAAUGCUAGUUCCACACUAAACAUACAUAUGUGUAUGCUGAUAAGGAU .(((((((.....)))))..-..........(((..((((((....((((...))))...))))))..)))(((((....)))))........((((((....)))))).......)).. ( -26.20) >DroSec_CAF1 38279 92 - 1 ACCAUGGUUUAGAGCCGUACUAAUAAUC----------------------------UAAUAAUCUUUAAGAGGAAUGCUAGUUCCUCACUAAACAUACAUAUGUGUUUGCUGAUAAGGAU ...(((((.....)))))..........----------------------------.....(((((((.(((((((....)))))))..((((((((....))))))))....))))))) ( -22.00) >DroSim_CAF1 38475 119 - 1 ACCAUGGUUUAGAGCCGUAC-AAUAUUUUAAUUUCGAGGAGUUUAAAGCCUCUGUGUAAUAAUCUUUAAGAGGAAUGCUAGUUCCUCACUAAACAUACAUAUGUGUGUGCUGAUAAGGAU ...(((((.....)))))..-..............((((.........)))).........(((((((.(((((((....))))))).(...((((((....))))))...).))))))) ( -27.90) >consensus ACCAUGGUUUAGAGCCGUAC_AAUAUUUUAAUUUCGAGGA_UUUAAAG_CUCUGU_UAAUAAUCUUUAAGAGGAAUGCUAGUUCCUCACUAAACAUACAUAUGUGUAUGCUGAUAAGGAU ...(((((.....)))))...........................................(((((((.(((((((....)))))))......((((((....))))))....))))))) (-21.45 = -21.57 + 0.12)

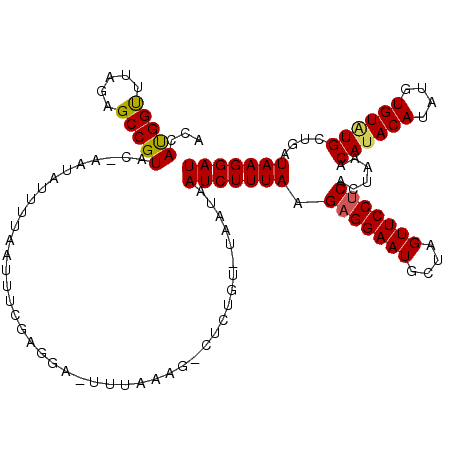

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:34 2006