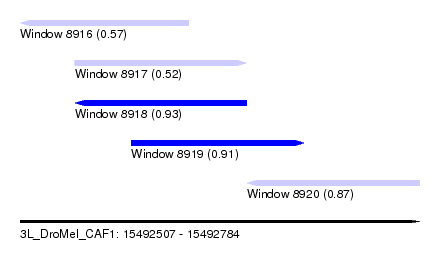
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,492,507 – 15,492,784 |
| Length | 277 |
| Max. P | 0.926623 |

| Location | 15,492,507 – 15,492,624 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.15 |
| Mean single sequence MFE | -39.81 |
| Consensus MFE | -29.75 |
| Energy contribution | -31.62 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15492507 117 - 23771897 CUGCGAUGGCGAUGAUUCCUACUACUGCAAUGGGGAUUGUCGUGAGGCCGUGGAGUGUUAUACCAAUGUGGUUACCACA---CCGGUUACCACAUUAAGACCCAUAGGAACCUCUACGGA ......(.((((..(((((((.........)))))))..)))).)..((((((((.(((.....((((((((.(((...---..))).))))))))....((....))))))))))))). ( -44.50) >DroSec_CAF1 33724 120 - 1 CUGCGAUGGCGAUGACUCCUACUACUGCAAUGGGGAUUGUCGUGAGCCCGUGGAGUGUCAUACCAAUGUGGUUACCACACCAUCGGUUACCACCUUAAGACCCAUAGGAACCUCUACGGA ..((....((((..(..((((.........))))..)..))))..))((((((((.((.........(((((.(((........))).))))).......((....)).)))))))))). ( -36.80) >DroSim_CAF1 35750 120 - 1 CUGCGAUGGCGAUGAUUCCUACUACUGCAAUGGGGAUUGUCGUGAGCCCGUGGAGUGUCAUACCAAUGUGGUAACCAGACCACCGGUUACCACCUUAAGACCCAUAGGAACCUCUACGGA ..((....((((..(((((((.........)))))))..))))..))((((((((.((.........(((((((((........))))))))).......((....)).)))))))))). ( -43.80) >DroYak_CAF1 116124 117 - 1 CUGCGAUGGCGAUGAUUCCUACUACUGCAAUGGGGAUUGCCGCGAGGCGGUCGAGUGUUACACCAAUGUGGUUACCACG---CCAGCUACCACUUUACGGCCUACAGUAGCCACUACGAA ((((..(.(((..((((((((.........))))))))..))).).))))(((((((((((......((((...))))(---((..............))).....)))).)))).))). ( -34.14) >consensus CUGCGAUGGCGAUGAUUCCUACUACUGCAAUGGGGAUUGUCGUGAGCCCGUGGAGUGUCAUACCAAUGUGGUUACCACA___CCGGUUACCACCUUAAGACCCAUAGGAACCUCUACGGA ......(.(((((((((((((.........))))))))))))).)..((((((((.((.........(((((.(((........))).))))).......((....)).)))))))))). (-29.75 = -31.62 + 1.88)

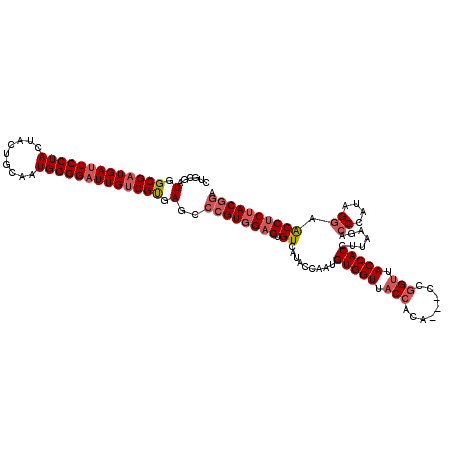

| Location | 15,492,545 – 15,492,664 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.35 |
| Mean single sequence MFE | -31.91 |
| Consensus MFE | -24.39 |
| Energy contribution | -24.20 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15492545 119 + 23771897 -UGUGGUAACCACAUUGGUAUAACACUCCACGGCCUCACGACAAUCCCCAUUGCAGUAGUAGGAAUCAUCGCCAUCGCAGUGAACAAACUCACUGCACUCCUGAUUCUCAUUGGUCACCU -.((((..(((.....)))........))))((((..((..((((....))))..))(((..((((((........(((((((......))))))).....))))))..))))))).... ( -29.52) >DroSec_CAF1 33764 120 + 1 GUGUGGUAACCACAUUGGUAUGACACUCCACGGGCUCACGACAAUCCCCAUUGCAGUAGUAGGAGUCAUCGCCAUCGCAGUGAACAAACUCACUGCACUCCUGAUUCUCAUUAGUCACCU ((((((...))))))(((((((((..(((....(((.((..((((....))))..))))).)))))))).))))..(((((((......))))))).....(((((......)))))... ( -33.60) >DroSim_CAF1 35790 120 + 1 GUCUGGUUACCACAUUGGUAUGACACUCCACGGGCUCACGACAAUCCCCAUUGCAGUAGUAGGAAUCAUCGCCAUCGCAGUGAACAAACUCACUGCACUCCUGAUUCUCAUUAGUCACCU ....(((((((.....)))).(((.......(((............))).......((((..((((((........(((((((......))))))).....))))))..)))))))))). ( -29.32) >DroYak_CAF1 116162 119 + 1 -CGUGGUAACCACAUUGGUGUAACACUCGACCGCCUCGCGGCAAUCCCCAUUGCAGUAGUAGGAAUCAUCGCCAUCGCAGUGAACGAAUUCACUGCACUCCUGGUUCUCGUUGGUCACCU -.(((...(((.....)))....)))..(((((((....)))...........(((.(((.((........))...((((((((....))))))))))).))).........)))).... ( -35.20) >consensus _UGUGGUAACCACAUUGGUAUAACACUCCACGGCCUCACGACAAUCCCCAUUGCAGUAGUAGGAAUCAUCGCCAUCGCAGUGAACAAACUCACUGCACUCCUGAUUCUCAUUAGUCACCU ....(((.(((.....)))....................(((...........(((.(((.((........))...(((((((......)))))))))).)))..........)))))). (-24.39 = -24.20 + -0.19)

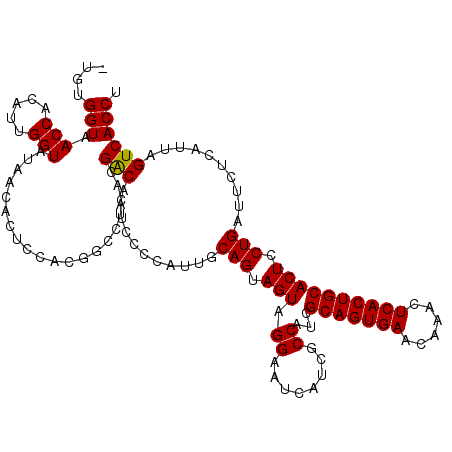
| Location | 15,492,545 – 15,492,664 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.35 |
| Mean single sequence MFE | -41.86 |
| Consensus MFE | -37.06 |
| Energy contribution | -37.94 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15492545 119 - 23771897 AGGUGACCAAUGAGAAUCAGGAGUGCAGUGAGUUUGUUCACUGCGAUGGCGAUGAUUCCUACUACUGCAAUGGGGAUUGUCGUGAGGCCGUGGAGUGUUAUACCAAUGUGGUUACCACA- .((((((((((((.(.((.((..(((((((((....))))))))).(.((((..(((((((.........)))))))..)))).)..))...)).).)))).......))))))))...- ( -42.11) >DroSec_CAF1 33764 120 - 1 AGGUGACUAAUGAGAAUCAGGAGUGCAGUGAGUUUGUUCACUGCGAUGGCGAUGACUCCUACUACUGCAAUGGGGAUUGUCGUGAGCCCGUGGAGUGUCAUACCAAUGUGGUUACCACAC .((((((((((((.(.((.((.((((((((((....))))))))..(.((((..(..((((.........))))..)..)))).)))))...)).).)))).......)))))))).... ( -40.01) >DroSim_CAF1 35790 120 - 1 AGGUGACUAAUGAGAAUCAGGAGUGCAGUGAGUUUGUUCACUGCGAUGGCGAUGAUUCCUACUACUGCAAUGGGGAUUGUCGUGAGCCCGUGGAGUGUCAUACCAAUGUGGUAACCAGAC .(((......(((.(.((.((.((((((((((....))))))))..(.((((..(((((((.........)))))))..)))).)))))...)).).)))((((.....))))))).... ( -39.80) >DroYak_CAF1 116162 119 - 1 AGGUGACCAACGAGAACCAGGAGUGCAGUGAAUUCGUUCACUGCGAUGGCGAUGAUUCCUACUACUGCAAUGGGGAUUGCCGCGAGGCGGUCGAGUGUUACACCAAUGUGGUUACCACG- .(((((((..........((((((((((((((....))))))))(....)....))))))......(((.(((.(((((((....)))))))..(.....).))).))))))))))...- ( -45.50) >consensus AGGUGACCAAUGAGAAUCAGGAGUGCAGUGAGUUUGUUCACUGCGAUGGCGAUGAUUCCUACUACUGCAAUGGGGAUUGUCGUGAGCCCGUGGAGUGUCAUACCAAUGUGGUUACCACA_ .((((((((((((.(.((.((..(((((((((....))))))))).(.(((((((((((((.........))))))))))))).)..))...)).).)))).......)))))))).... (-37.06 = -37.94 + 0.87)

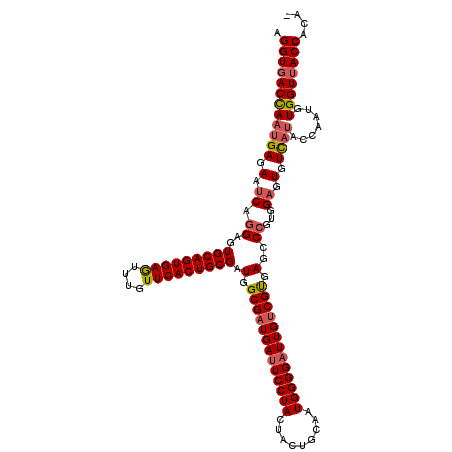
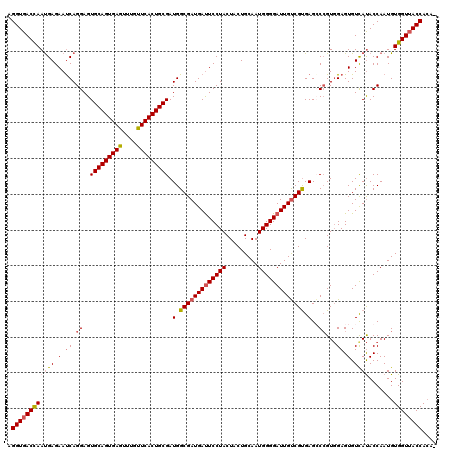
| Location | 15,492,584 – 15,492,704 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.56 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -18.41 |
| Energy contribution | -17.59 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15492584 120 + 23771897 ACAAUCCCCAUUGCAGUAGUAGGAAUCAUCGCCAUCGCAGUGAACAAACUCACUGCACUCCUGAUUCUCAUUGGUCACCUUAUUGAUGAUCACACCUUUGCAUUCCUCGAGAAAAACCGC ...........(((((..((.(..((((((......(((((((......))))))).....(((((......))))).......))))))..)))..))))).................. ( -27.20) >DroSec_CAF1 33804 120 + 1 ACAAUCCCCAUUGCAGUAGUAGGAGUCAUCGCCAUCGCAGUGAACAAACUCACUGCACUCCUGAUUCUCAUUAGUCACCUUAUUGAUGAUCACACCCUUGCAUUCCUCGAGAAAAACCGC ...........(((((..((.(..((((((......(((((((......))))))).....(((((......))))).......))))))..)))..))))).................. ( -26.80) >DroSim_CAF1 35830 120 + 1 ACAAUCCCCAUUGCAGUAGUAGGAAUCAUCGCCAUCGCAGUGAACAAACUCACUGCACUCCUGAUUCUCAUUAGUCACCUUANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN ...........((.(.((((..((((((........(((((((......))))))).....))))))..)))).)))........................................... ( -18.62) >DroYak_CAF1 116201 120 + 1 GCAAUCCCCAUUGCAGUAGUAGGAAUCAUCGCCAUCGCAGUGAACGAAUUCACUGCACUCCUGGUUCUCGUUGGUCACCUUAUUGAUGAUCACACCUUUGCAUUCCUCGAGGAAAACCGC ((.........(((((..((.(..((((((......((((((((....))))))))......(((((.....))..))).....))))))..)))..)))))((((....))))....)) ( -31.30) >consensus ACAAUCCCCAUUGCAGUAGUAGGAAUCAUCGCCAUCGCAGUGAACAAACUCACUGCACUCCUGAUUCUCAUUAGUCACCUUAUUGAUGAUCACACCUUUGCAUUCCUCGAGAAAAACCGC ...........((.(.((((..((((((........(((((((......))))))).....))))))..)))).)))........................................... (-18.41 = -17.59 + -0.81)

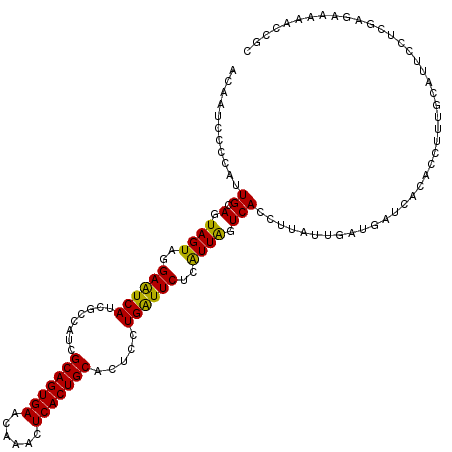
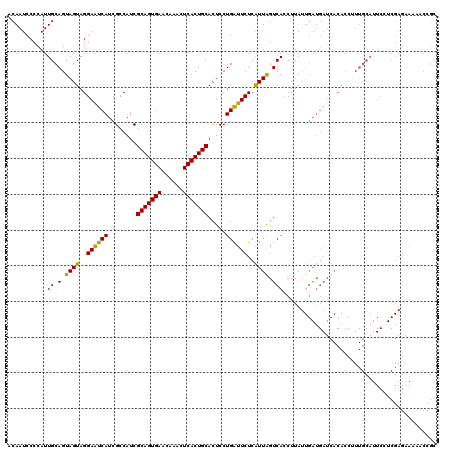
| Location | 15,492,664 – 15,492,784 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.30 |
| Mean single sequence MFE | -41.03 |
| Consensus MFE | -38.51 |
| Energy contribution | -38.30 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15492664 120 - 23771897 UGCAGAAAGCCUAUUGAAUGUGGCAAUUGUUGAUAGUCUUGGCCUGCCUGUUACCUGGCUACCUCUGGGUGCAGGUGCAGGCGGUUUUUCUCGAGGAAUGCAAAGGUGUGAUCAUCAAUA ........(((..........)))...(((((((.(((...((((((((((.(((((.(.(((....))))))))))))))).((.((((....)))).))..))))..))).))))))) ( -40.20) >DroSec_CAF1 33884 118 - 1 U--UGAAAGCCUAUUGAAUGUGGCAAUUGUUGAUAGUCUUGGCCUGCCUGUUACCUGGCUACCUAUGGUUGCAGGUGCAGGCGGUUUUUCUCGAGGAAUGCAAGGGUGUGAUCAUCAAUA .--.....(((..........)))...(((((((.(((...((((((((((.(((((.(.(((...))).)))))))))))).((.((((....)))).))..))))..))).))))))) ( -38.90) >DroYak_CAF1 116281 118 - 1 U--UGAAAGCCUAUUGAAUGUGGCAAUUGUUGUUAGUCUUGGCCUGCCUGCUACCUGGUUGCCUCCGGGUGCAGGUGCAGGCGGUUUUCCUCGAGGAAUGCAAAGGUGUGAUCAUCAAUA .--........((((((..((.(((....((((...(((((((((((((((.((((((......))))))))))..))))))((....)).)))))...))))...))).))..)))))) ( -44.00) >consensus U__UGAAAGCCUAUUGAAUGUGGCAAUUGUUGAUAGUCUUGGCCUGCCUGUUACCUGGCUACCUCUGGGUGCAGGUGCAGGCGGUUUUUCUCGAGGAAUGCAAAGGUGUGAUCAUCAAUA ........(((..........)))...(((((((.(((...((((((((((.(((((..((((....))))))))))))))).((.((((....)))).))..))))..))).))))))) (-38.51 = -38.30 + -0.21)

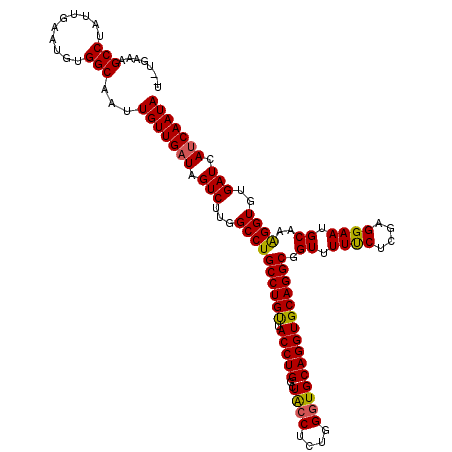
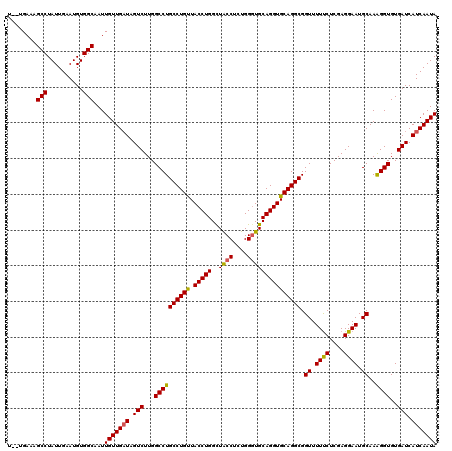
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:27 2006