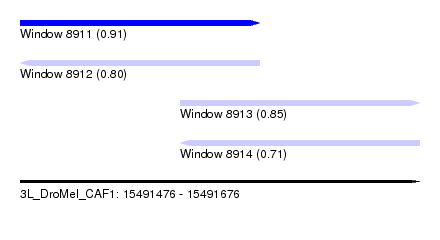
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,491,476 – 15,491,676 |
| Length | 200 |
| Max. P | 0.906749 |

| Location | 15,491,476 – 15,491,596 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -38.68 |
| Consensus MFE | -34.00 |
| Energy contribution | -35.12 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15491476 120 + 23771897 UCGCAUUUUCCGGUGAGUGAAUUAAAGUGCAAACUGGUGGCACAACUCAUGGGAAGUGCCACACCGCGAUAGCAAAUGUAAUAAUCUGAACAGGAGUUCUGAUUUGGCAGCAAUGCGAUA (((((((.((((((...........(((....))).(((((((..((....))..))))))))))).))..((...(((...((((.((((....)))).))))..)))))))))))).. ( -38.70) >DroSec_CAF1 32696 120 + 1 UCGCAUUUUCCGGUGCGUGAAUUAAAAUGCAAACUGGUGGCACAGCUCAUGGGAAGUGCCACUCCGCGAUAGCAAAUGUAAUAAUCUGAACAGGAGUUCUGAUUUGCCAGCAAUGCGAUA (((((((.((((((((((........)))))....((((((((..((....))..))))))))))).))..((....((((..(((.((((....)))).)))))))..))))))))).. ( -38.70) >DroSim_CAF1 34722 120 + 1 UCGCAUUUUCCGGUGCGUGAAUUAAAAUGCAAACUGGUGGCACAGCUCAUGGGAAGUGCCACUCCGCGAUAGCAAAUGUAAUAAUCUGAACAGGAGUUCUGAUUUGCCAGCAAUGCGAUA (((((((.((((((((((........)))))....((((((((..((....))..))))))))))).))..((....((((..(((.((((....)))).)))))))..))))))))).. ( -38.70) >DroYak_CAF1 115096 120 + 1 UCGCAUUUUCCGGUGCGGGAAUUAAAGUGCAAACUGGUGGCACAACUCAUGGGGAGUGCCUCGCCGCGAUAGCAGAUGUAAUAAUCUGAACAGGAGUUCUGAUUUGGCAGCAAUGCGAUA ((((((((((((...))))))......(((......(.(((((..((....))..))))).)(((((....)).........((((.((((....)))).)))).))).))))))))).. ( -38.60) >consensus UCGCAUUUUCCGGUGCGUGAAUUAAAAUGCAAACUGGUGGCACAACUCAUGGGAAGUGCCACUCCGCGAUAGCAAAUGUAAUAAUCUGAACAGGAGUUCUGAUUUGCCAGCAAUGCGAUA (((((((.((((((((((........))))).....(((((((..((....))..))))))).))).))..((...(((...((((.((((....)))).))))..)))))))))))).. (-34.00 = -35.12 + 1.12)

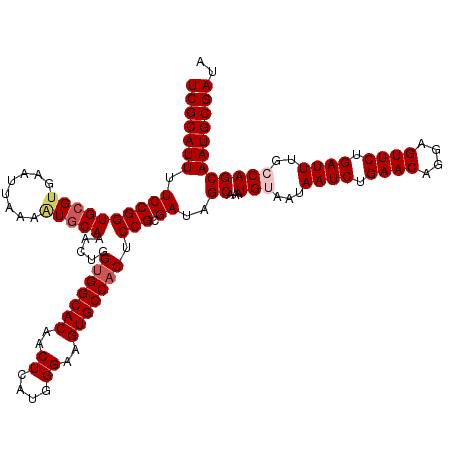
| Location | 15,491,476 – 15,491,596 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -33.78 |
| Consensus MFE | -29.15 |
| Energy contribution | -29.90 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15491476 120 - 23771897 UAUCGCAUUGCUGCCAAAUCAGAACUCCUGUUCAGAUUAUUACAUUUGCUAUCGCGGUGUGGCACUUCCCAUGAGUUGUGCCACCAGUUUGCACUUUAAUUCACUCACCGGAAAAUGCGA ..(((((((..((((.((((.((((....)))).))))...(((((.((....))))))))))).((((..(((((.((((.........))))........)))))..))))))))))) ( -33.60) >DroSec_CAF1 32696 120 - 1 UAUCGCAUUGCUGGCAAAUCAGAACUCCUGUUCAGAUUAUUACAUUUGCUAUCGCGGAGUGGCACUUCCCAUGAGCUGUGCCACCAGUUUGCAUUUUAAUUCACGCACCGGAAAAUGCGA ..(((((((.((((..((((.((((....)))).))))...............((((((((((((............)))))))..(....)........)).))).))))..))))))) ( -34.10) >DroSim_CAF1 34722 120 - 1 UAUCGCAUUGCUGGCAAAUCAGAACUCCUGUUCAGAUUAUUACAUUUGCUAUCGCGGAGUGGCACUUCCCAUGAGCUGUGCCACCAGUUUGCAUUUUAAUUCACGCACCGGAAAAUGCGA ..(((((((.((((..((((.((((....)))).))))...............((((((((((((............)))))))..(....)........)).))).))))..))))))) ( -34.10) >DroYak_CAF1 115096 120 - 1 UAUCGCAUUGCUGCCAAAUCAGAACUCCUGUUCAGAUUAUUACAUCUGCUAUCGCGGCGAGGCACUCCCCAUGAGUUGUGCCACCAGUUUGCACUUUAAUUCCCGCACCGGAAAAUGCGA ..(((((((((((((.((((.((((....)))).)))).........((....)))))).(((((..(......)..)))))........)).......((((......))))))))))) ( -33.30) >consensus UAUCGCAUUGCUGCCAAAUCAGAACUCCUGUUCAGAUUAUUACAUUUGCUAUCGCGGAGUGGCACUUCCCAUGAGCUGUGCCACCAGUUUGCACUUUAAUUCACGCACCGGAAAAUGCGA ..(((((((.((((..((((.((((....)))).))))........(((....((.(.(((((((............)))))))).))..)))..............))))..))))))) (-29.15 = -29.90 + 0.75)



| Location | 15,491,556 – 15,491,676 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.39 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -33.71 |
| Energy contribution | -34.77 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15491556 120 + 23771897 AUAAUCUGAACAGGAGUUCUGAUUUGGCAGCAAUGCGAUACGACUCUGAUUAUCCAGUUGUGGGCACACAUUGGUCACAAUAAUCUCUAUAGCAGGCGAUACGGAAGUCGUACUGCUGGC ..((((.((((....)))).)))).....((...(((.(((((((((((((..((.((((((((.....((((....))))....)))))))).)).))).))).))))))).)))..)) ( -33.00) >DroSec_CAF1 32776 120 + 1 AUAAUCUGAACAGGAGUUCUGAUUUGCCAGCAAUGCGAUACGACUCUGAUUAUCCAGUUGUGGGCACACAUUGGUCACAAUAAUCUCUAUAGCAGGCGAUACGGAAGUCGCAUUGCUGGC ..((((.((((....)))).)))).((((((((((((((.....(((((((..((.((((((((.....((((....))))....)))))))).)).))).)))).)))))))))))))) ( -42.70) >DroSim_CAF1 34802 120 + 1 AUAAUCUGAACAGGAGUUCUGAUUUGCCAGCAAUGCGAUACGACUCUGAUUAUCCAGUUGUGGGCACACAUUGGUCACAAUAAUGUCUAUAGCAGGCGAUACGGAAGUCGCAUUGCUGGC ..((((.((((....)))).)))).((((((((((((((.....(((((((..((.((((((((((...((((....))))..)))))))))).)).))).)))).)))))))))))))) ( -47.90) >DroYak_CAF1 115176 120 + 1 AUAAUCUGAACAGGAGUUCUGAUUUGGCAGCAAUGCGAUACGACUCUGAUUAUCCAGUUGCGGGCACACAUUGGUCACAAUAAUCUCUAUGCCAGGCGAUACGGAAGUCGUAUUGGUGGC ..((((.((((....)))).))))..(((....)))(((((((((.......(((.(((((.((((...((((....))))........))))..)))))..))))))))))))...... ( -31.91) >consensus AUAAUCUGAACAGGAGUUCUGAUUUGCCAGCAAUGCGAUACGACUCUGAUUAUCCAGUUGUGGGCACACAUUGGUCACAAUAAUCUCUAUAGCAGGCGAUACGGAAGUCGCAUUGCUGGC ..((((.((((....)))).)))).((((((((((((((.....(((((((..((.((((((((.....((((....))))....)))))))).)).))).)))).)))))))))))))) (-33.71 = -34.77 + 1.06)



| Location | 15,491,556 – 15,491,676 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.39 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -30.16 |
| Energy contribution | -30.85 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15491556 120 - 23771897 GCCAGCAGUACGACUUCCGUAUCGCCUGCUAUAGAGAUUAUUGUGACCAAUGUGUGCCCACAACUGGAUAAUCAGAGUCGUAUCGCAUUGCUGCCAAAUCAGAACUCCUGUUCAGAUUAU (.((((((((((((((..(((.....)))......((((((.....(((.((((....))))..))))))))).))))))))).....))))).).((((.((((....)))).)))).. ( -29.70) >DroSec_CAF1 32776 120 - 1 GCCAGCAAUGCGACUUCCGUAUCGCCUGCUAUAGAGAUUAUUGUGACCAAUGUGUGCCCACAACUGGAUAAUCAGAGUCGUAUCGCAUUGCUGGCAAAUCAGAACUCCUGUUCAGAUUAU (((((((((((((((...(((.....)))...)).((((((.....(((.((((....))))..))))))))).........))))))))))))).((((.((((....)))).)))).. ( -39.40) >DroSim_CAF1 34802 120 - 1 GCCAGCAAUGCGACUUCCGUAUCGCCUGCUAUAGACAUUAUUGUGACCAAUGUGUGCCCACAACUGGAUAAUCAGAGUCGUAUCGCAUUGCUGGCAAAUCAGAACUCCUGUUCAGAUUAU (((((((((((((.....(((.....)))....(((....(((((..((.....))..)))))(((......))).)))...))))))))))))).((((.((((....)))).)))).. ( -40.40) >DroYak_CAF1 115176 120 - 1 GCCACCAAUACGACUUCCGUAUCGCCUGGCAUAGAGAUUAUUGUGACCAAUGUGUGCCCGCAACUGGAUAAUCAGAGUCGUAUCGCAUUGCUGCCAAAUCAGAACUCCUGUUCAGAUUAU .......(((((((((...((((...((((((((......))))).))).((((....))))....))))....))))))))).(((....)))..((((.((((....)))).)))).. ( -27.70) >consensus GCCAGCAAUACGACUUCCGUAUCGCCUGCUAUAGAGAUUAUUGUGACCAAUGUGUGCCCACAACUGGAUAAUCAGAGUCGUAUCGCAUUGCUGCCAAAUCAGAACUCCUGUUCAGAUUAU (((((((((((((......(((.(((((.(((((......))))).(((.((((....))))..))).....))).)).)))))))))))))))).((((.((((....)))).)))).. (-30.16 = -30.85 + 0.69)

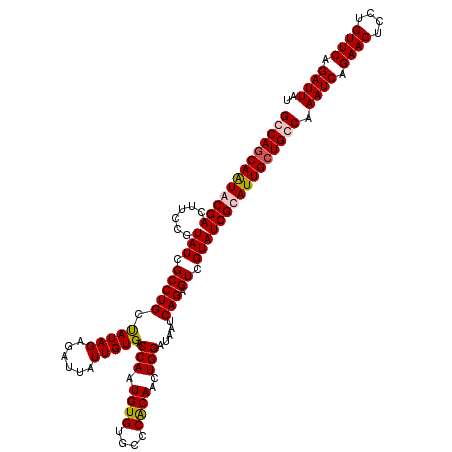
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:21 2006