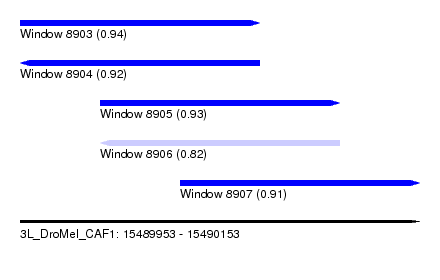
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,489,953 – 15,490,153 |
| Length | 200 |
| Max. P | 0.940506 |

| Location | 15,489,953 – 15,490,073 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -40.24 |
| Consensus MFE | -31.95 |
| Energy contribution | -34.64 |
| Covariance contribution | 2.69 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15489953 120 + 23771897 UCCAUAGUAGCAAAUAUAGUACUUUCUGCAACUACUCGAGCUGCCGACAUAUACAAUCCUGUCGCUGGAUUGUUGUUGCUCCACCAACGUGGAGCAAUAAGUGGCGGCAUCCGUGACCAU ....((((.(((..............))).)))).....((((((.((.....((((((.......))))))((((((((((((....))))))))))))))))))))............ ( -41.84) >DroSec_CAF1 31157 120 + 1 UCCAUAGUAGCAAAUAUAGUAAUUUCUGCAACUGCUCGAGCUGCCGACAUAUACAAUCCUGUCGCUGGAUUGUUGUUGCUCCACUAACGUGGAGCAAUAGGUGGCGGCAUCCGUGACCAU ..(((.((((.((((......))))))))..........(((((((.(....(((((((.......)))))))(((((((((((....)))))))))))).)))))))....)))..... ( -45.10) >DroSim_CAF1 33183 120 + 1 UCCAUAGUAGCAAAUAUAGUACUUUCUGCAACUACUCGAGCUGCCGACAUAUACAAUCCUGUCGCUGGAUUGUUGUUGCUCCACUAACGUGGAGCAAUAGGUGGCGGCAUCCGUGACCAU ....((((.(((..............))).)))).....(((((((.(....(((((((.......)))))))(((((((((((....)))))))))))).)))))))............ ( -44.14) >DroYak_CAF1 113537 120 + 1 UCCAUAGUAGCAAAUAUAGUAUUGUCUGCAACUACUAGAACUACCGACAUAAACCAGUCUGUCGCUGGAUUGUGGUUGGCCCAUUAACGUGGAGCAAAACGUGGCUGCAUCCGUGACCAU ......(((((.....((((((((....))).)))))....(((((((((((.(((((.....))))).)))).))))((((((....)))).)).....))))))))............ ( -29.90) >consensus UCCAUAGUAGCAAAUAUAGUACUUUCUGCAACUACUCGAGCUGCCGACAUAUACAAUCCUGUCGCUGGAUUGUUGUUGCUCCACUAACGUGGAGCAAUAGGUGGCGGCAUCCGUGACCAU ....((((.(((..............))).)))).....((((((.((....(((((((.......)))))))(((((((((((....))))))))))).))))))))............ (-31.95 = -34.64 + 2.69)

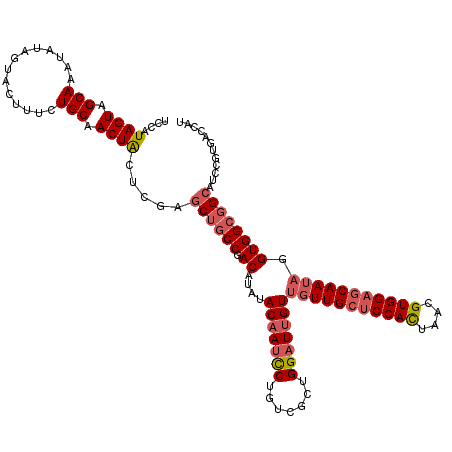

| Location | 15,489,953 – 15,490,073 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -39.05 |
| Consensus MFE | -32.34 |
| Energy contribution | -33.27 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15489953 120 - 23771897 AUGGUCACGGAUGCCGCCACUUAUUGCUCCACGUUGGUGGAGCAACAACAAUCCAGCGACAGGAUUGUAUAUGUCGGCAGCUCGAGUAGUUGCAGAAAGUACUAUAUUUGCUACUAUGGA .......(((.(((((.((....(((((((((....)))))))))..(((((((.......)))))))...)).)))))..))).(((((.(((((.(......).))))).)))))... ( -42.70) >DroSec_CAF1 31157 120 - 1 AUGGUCACGGAUGCCGCCACCUAUUGCUCCACGUUAGUGGAGCAACAACAAUCCAGCGACAGGAUUGUAUAUGUCGGCAGCUCGAGCAGUUGCAGAAAUUACUAUAUUUGCUACUAUGGA (((((..(((.(((((.((....(((((((((....)))))))))..(((((((.......)))))))...)).)))))..)))(((((.((.((......)).)).))))))))))... ( -41.50) >DroSim_CAF1 33183 120 - 1 AUGGUCACGGAUGCCGCCACCUAUUGCUCCACGUUAGUGGAGCAACAACAAUCCAGCGACAGGAUUGUAUAUGUCGGCAGCUCGAGUAGUUGCAGAAAGUACUAUAUUUGCUACUAUGGA .......(((.(((((.((....(((((((((....)))))))))..(((((((.......)))))))...)).)))))..))).(((((.(((((.(......).))))).)))))... ( -43.30) >DroYak_CAF1 113537 120 - 1 AUGGUCACGGAUGCAGCCACGUUUUGCUCCACGUUAAUGGGCCAACCACAAUCCAGCGACAGACUGGUUUAUGUCGGUAGUUCUAGUAGUUGCAGACAAUACUAUAUUUGCUACUAUGGA .(((((..(((.((((.......))))))).((....)))))))((((((..(((((....).))))....))).)))...((((.((((.(((((..........))))).)))))))) ( -28.70) >consensus AUGGUCACGGAUGCCGCCACCUAUUGCUCCACGUUAGUGGAGCAACAACAAUCCAGCGACAGGAUUGUAUAUGUCGGCAGCUCGAGUAGUUGCAGAAAGUACUAUAUUUGCUACUAUGGA .........((.((.(((.....(((((((((....)))))))))..(((((((.......))))))).......))).))))..(((((.(((((..........))))).)))))... (-32.34 = -33.27 + 0.94)

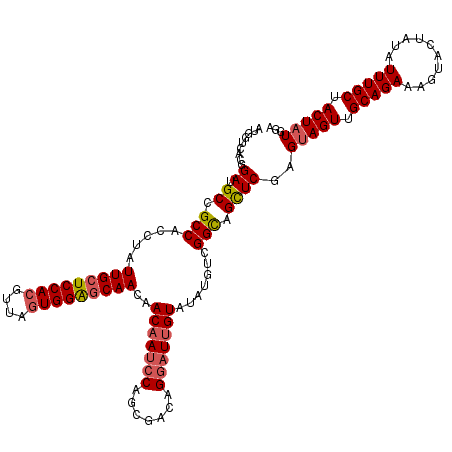

| Location | 15,489,993 – 15,490,113 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -43.97 |
| Consensus MFE | -34.46 |
| Energy contribution | -36.52 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.54 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15489993 120 + 23771897 CUGCCGACAUAUACAAUCCUGUCGCUGGAUUGUUGUUGCUCCACCAACGUGGAGCAAUAAGUGGCGGCAUCCGUGACCAUGUUAUUCGGAUCUCCAUCACCAUUUCCGCCAUCGAAUGGA .(((((.(((...((((((.......))))))((((((((((((....))))))))))))))).)))))((((((((...))))..)))).........((((((........)))))). ( -43.80) >DroSec_CAF1 31197 120 + 1 CUGCCGACAUAUACAAUCCUGUCGCUGGAUUGUUGUUGCUCCACUAACGUGGAGCAAUAGGUGGCGGCAUCCGUGACCAUGUUAUUUGGAUCUCCAUCACCAUUUCCGCCAUCGAAUGGA ...(((......(((((((.......)))))))(((((((((((....)))))))))))((((((((.....((((..........(((....))))))).....))))))))...))). ( -48.20) >DroSim_CAF1 33223 120 + 1 CUGCCGACAUAUACAAUCCUGUCGCUGGAUUGUUGUUGCUCCACUAACGUGGAGCAAUAGGUGGCGGCAUCCGUGACCAUGUUAUUUGGAUCUCCAUCACCAUUUCCGCCAUCGAAUGGA ...(((......(((((((.......)))))))(((((((((((....)))))))))))((((((((.....((((..........(((....))))))).....))))))))...))). ( -48.20) >DroYak_CAF1 113577 120 + 1 CUACCGACAUAAACCAGUCUGUCGCUGGAUUGUGGUUGGCCCAUUAACGUGGAGCAAAACGUGGCUGCAUCCGUGACCAUGUCAUUAGGAUCUCCAUCACCGUUUCCACCAUCAAAUGGA ...(((((((((.(((((.....))))).)))).))))).(((((...(((((((...((((((..((....))..)))))).....((....))......).)))))).....))))). ( -35.70) >consensus CUGCCGACAUAUACAAUCCUGUCGCUGGAUUGUUGUUGCUCCACUAACGUGGAGCAAUAGGUGGCGGCAUCCGUGACCAUGUUAUUUGGAUCUCCAUCACCAUUUCCGCCAUCGAAUGGA .(((((.(((..(((((((.......)))))))(((((((((((....))))))))))).))).)))))((((((((...)))))..))).........((((((........)))))). (-34.46 = -36.52 + 2.06)

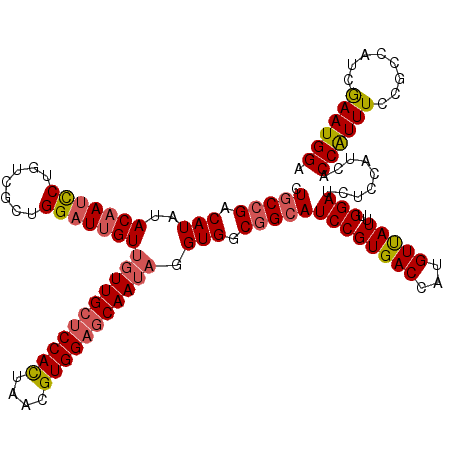

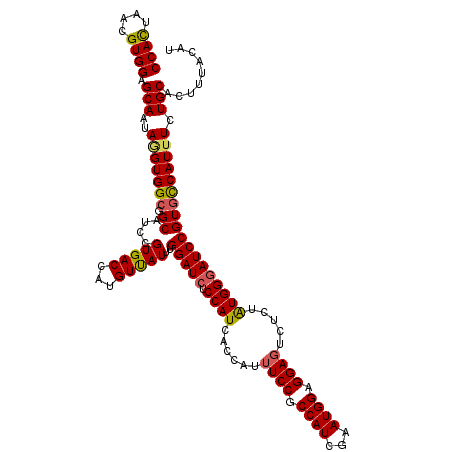
| Location | 15,489,993 – 15,490,113 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -39.80 |
| Consensus MFE | -34.72 |
| Energy contribution | -35.48 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15489993 120 - 23771897 UCCAUUCGAUGGCGGAAAUGGUGAUGGAGAUCCGAAUAACAUGGUCACGGAUGCCGCCACUUAUUGCUCCACGUUGGUGGAGCAACAACAAUCCAGCGACAGGAUUGUAUAUGUCGGCAG .....((((((((((...(.(((((((....))(.....)...))))).)...))))).....(((((((((....)))))))))..(((((((.......)))))))....)))))... ( -42.80) >DroSec_CAF1 31197 120 - 1 UCCAUUCGAUGGCGGAAAUGGUGAUGGAGAUCCAAAUAACAUGGUCACGGAUGCCGCCACCUAUUGCUCCACGUUAGUGGAGCAACAACAAUCCAGCGACAGGAUUGUAUAUGUCGGCAG .....((((((((((...(.(((((((....)))..........)))).)...))))).....(((((((((....)))))))))..(((((((.......)))))))....)))))... ( -43.00) >DroSim_CAF1 33223 120 - 1 UCCAUUCGAUGGCGGAAAUGGUGAUGGAGAUCCAAAUAACAUGGUCACGGAUGCCGCCACCUAUUGCUCCACGUUAGUGGAGCAACAACAAUCCAGCGACAGGAUUGUAUAUGUCGGCAG .....((((((((((...(.(((((((....)))..........)))).)...))))).....(((((((((....)))))))))..(((((((.......)))))))....)))))... ( -43.00) >DroYak_CAF1 113577 120 - 1 UCCAUUUGAUGGUGGAAACGGUGAUGGAGAUCCUAAUGACAUGGUCACGGAUGCAGCCACGUUUUGCUCCACGUUAAUGGGCCAACCACAAUCCAGCGACAGACUGGUUUAUGUCGGUAG ((((((..((.((....)).))))))))....(((.((((((((....((.((..(((.((((..((.....)).))))))))).)).....(((((....).)))).)))))))).))) ( -30.40) >consensus UCCAUUCGAUGGCGGAAAUGGUGAUGGAGAUCCAAAUAACAUGGUCACGGAUGCCGCCACCUAUUGCUCCACGUUAGUGGAGCAACAACAAUCCAGCGACAGGAUUGUAUAUGUCGGCAG .....((((((((((...(.(((((((....))..........))))).)...))))).....(((((((((....)))))))))..(((((((.......)))))))....)))))... (-34.72 = -35.48 + 0.75)

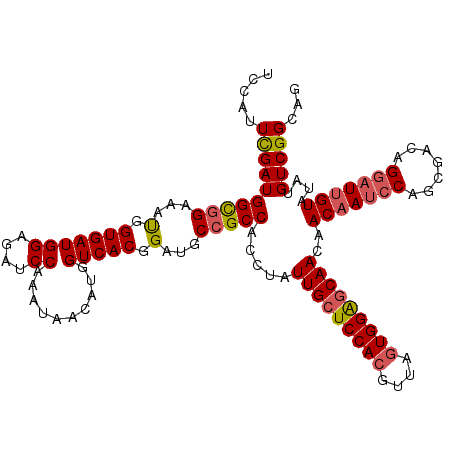

| Location | 15,490,033 – 15,490,153 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -36.75 |
| Consensus MFE | -32.66 |
| Energy contribution | -33.73 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914288 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15490033 120 + 23771897 CCACCAACGUGGAGCAAUAAGUGGCGGCAUCCGUGACCAUGUUAUUCGGAUCUCCAUCACCAUUUCCGCCAUCGAAUGGAGGAGUCUCUAUGGGAUCCGUUUCAUUUCUGCACUUUACAU ((((....)))).(((....((.((((...)))).)).(((.....((((((.((((......((((.((((...)))).)))).....))))))))))...)))...)))......... ( -35.00) >DroSec_CAF1 31237 120 + 1 CCACUAACGUGGAGCAAUAGGUGGCGGCAUCCGUGACCAUGUUAUUUGGAUCUCCAUCACCAUUUCCGCCAUCGAAUGGAGGAGUCUCUAUGGGAUCCGUGUCAUUUCUGCACUUUACAU ((((....)))).(((...(((.((((...)))).)))(((.....((((((.((((......((((.((((...)))).)))).....))))))))))...)))...)))......... ( -40.00) >DroSim_CAF1 33263 120 + 1 CCACUAACGUGGAGCAAUAGGUGGCGGCAUCCGUGACCAUGUUAUUUGGAUCUCCAUCACCAUUUCCGCCAUCGAAUGGAGGAGUCUCUGUGGGAUCCGUGCCAUUUCUGCACUUUACAU ((((....))))((((...(((.((((...)))).))).))))....(((((.((((......((((.((((...)))).)))).....)))))))))((((.......))))....... ( -41.50) >DroYak_CAF1 113617 120 + 1 CCAUUAACGUGGAGCAAAACGUGGCUGCAUCCGUGACCAUGUCAUUAGGAUCUCCAUCACCGUUUCCACCAUCAAAUGGAGGAUUCUCCAUGGCAUCCGUGCCAUUUCUGCACUUUACAU ........(((((((...((((((..((....))..)))))).....((....))......).))))))......((((((....))))))((((....))))................. ( -30.50) >consensus CCACUAACGUGGAGCAAUAGGUGGCGGCAUCCGUGACCAUGUUAUUUGGAUCUCCAUCACCAUUUCCGCCAUCGAAUGGAGGAGUCUCUAUGGGAUCCGUGCCAUUUCUGCACUUUACAU ((((....)))).(((..(((((((.((....(((((...)))))..(((((.((((......((((.((((...)))).)))).....)))))))))))))))))).)))......... (-32.66 = -33.73 + 1.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:14 2006