
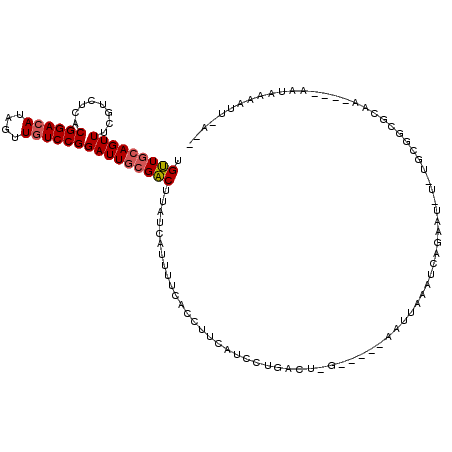
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,464,270 – 15,464,441 |
| Length | 171 |
| Max. P | 0.887737 |

| Location | 15,464,270 – 15,464,380 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.29 |
| Mean single sequence MFE | -21.56 |
| Consensus MFE | -13.98 |
| Energy contribution | -14.53 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15464270 110 - 23771897 UGUUGCAGUUUGGUUUCUCGGACAUAGUUAUCCGGAUUGCGACUAAUCAUUUUCACCUUCAUCCUGAUU-G-----CAUUUAUUAAGCUUUUCUGCGGCGUAA----AAUAAAAUUAAUC .(((((((...(((((.(((((........)))))..(((((....(((...............)))))-)-----))......)))))...)))))))....----............. ( -19.96) >DroWil_CAF1 219214 106 - 1 UGUUGCAGUUUCGUCUCACGGACAUAAUUGUCCGGAUUGCGACUUAACAUUUUCACCUUCAUUUUGCCUAGCGUUCAAUUAAAUCAAAAU---------ACA-----AAUAUAAUUUACA .(((((((((........((((((....)))))))))))))))...............................................---------...-----............. ( -17.60) >DroAna_CAF1 332496 110 - 1 UGCUCCAGUUUUGUCUCCCGGACAUAGUUGUCCGGAUUACGGCUUAUCAUUUUCACCUUCAUCCUGGCA-G-----AAUUAAAUCAGUAUCUUUGCGGCGCAAUUAUCAUAAAAUU---- (((.(((((((((((..(((((((....))))))).....((.............))........))))-)-----))))......(((....))))).)))..............---- ( -27.12) >consensus UGUUGCAGUUUCGUCUCACGGACAUAGUUGUCCGGAUUGCGACUUAUCAUUUUCACCUUCAUCCUGACU_G_____AAUUAAAUCAGAAU_U_UGCGGCGCAA____AAUAAAAUU_A__ .(((((((((........((((((....)))))))))))))))............................................................................. (-13.98 = -14.53 + 0.56)


| Location | 15,464,306 – 15,464,409 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.22 |
| Mean single sequence MFE | -23.36 |
| Consensus MFE | -14.86 |
| Energy contribution | -15.53 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15464306 103 - 23771897 UUUC-UUCG----------UAUAUGAACUUACUUUUGUGCUGUUGCAGUUUGGUUUCUCGGACAUAGUUAUCCGGAUUGCGACUAAUCAUUUUCACCUUCAUCCUGAUU-G-----CAUU ....-...(----------((........)))....((((.((((((((((((...((.......))....)))))))))))).(((((...............)))))-)-----))). ( -19.06) >DroWil_CAF1 219240 119 - 1 UGUG-UCUGGAUGGAGCUAUCUAUAUAGUUACCUUUGUGUUGUUGCAGUUUCGUCUCACGGACAUAAUUGUCCGGAUUGCGACUUAACAUUUUCACCUUCAUUUUGCCUAGCGUUCAAUU ...(-(..((..((((((((....)))))).))..(((...(((((((((........((((((....)))))))))))))))...))).................))..))........ ( -28.70) >DroAna_CAF1 332532 100 - 1 GUGUUAUUG--------------UCUACUCACUUUUGUGAUGCUCCAGUUUUGUCUCCCGGACAUAGUUGUCCGGAUUACGGCUUAUCAUUUUCACCUUCAUCCUGGCA-G-----AAUU .........--------------.....((((....))))......(((((((((..(((((((....))))))).....((.............))........))))-)-----)))) ( -22.32) >consensus UUUC_UUUG__________U_UAUAUACUUACUUUUGUGAUGUUGCAGUUUCGUCUCACGGACAUAGUUGUCCGGAUUGCGACUUAUCAUUUUCACCUUCAUCCUGACU_G_____AAUU ....................................((((((((((((((........((((((....)))))))))))))))..))))).............................. (-14.86 = -15.53 + 0.67)


| Location | 15,464,340 – 15,464,441 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 60.41 |
| Mean single sequence MFE | -20.77 |
| Consensus MFE | -10.46 |
| Energy contribution | -10.13 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15464340 101 + 23771897 GCAAUCCGGAUAACUAUGUCCGAGAAACCAAACUGCAACAGCACAAAAGUAAGUUCAUAUA----------CGAA-GAAAGACCAAAGUAUUAUAAGAA------UGCCAC--UUUCAGU ....((((((((....)))))).))........(((....))).....(((........))----------)...-(((((......(((((.....))------)))..)--))))... ( -13.70) >DroWil_CAF1 219280 113 + 1 GCAAUCCGGACAAUUAUGUCCGUGAGACGAAACUGCAACAACACAAAGGUAACUAUAUAGAUAGCUCCAUCCAGA-CACAAACAAAAG----ACAUGAAUUUCCAUUCAAU--UUGCAGU ....((((((((....)))))).))......(((((((.........((...((((....))))..)).......-............----...(((((....)))))..--))))))) ( -22.50) >DroAna_CAF1 332566 100 + 1 GUAAUCCGGACAACUAUGUCCGGGAGACAAAACUGGAGCAUCACAAAAGUGAGUAGA--------------CAAUAACACGACAAUACGAUUCUGAGGC------UCUUAUUUUUUCAGU ....((((((((....)))))))).((.((((..(((((.((((....)))).....--------------........((......))........))------)))..)))).))... ( -26.10) >consensus GCAAUCCGGACAACUAUGUCCGAGAGACAAAACUGCAACAACACAAAAGUAAGUACAUA_A__________CAAA_AAAAGACAAAAG_AUUAUAAGAA______UCCAAU__UUUCAGU ....((((((((....)))))).))......(((((((...........................................................................))))))) (-10.46 = -10.13 + -0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:03 2006