
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,616,961 – 1,617,062 |
| Length | 101 |
| Max. P | 0.569203 |

| Location | 1,616,961 – 1,617,062 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.35 |
| Mean single sequence MFE | -38.60 |
| Consensus MFE | -20.07 |
| Energy contribution | -21.27 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
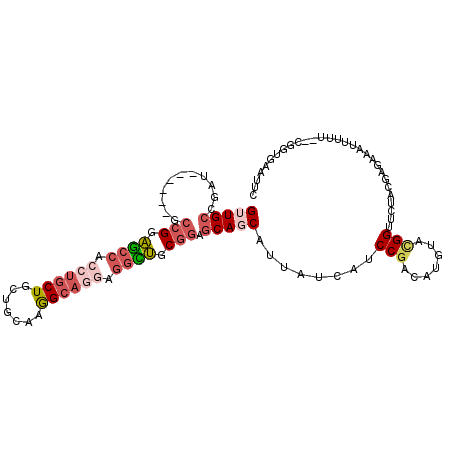
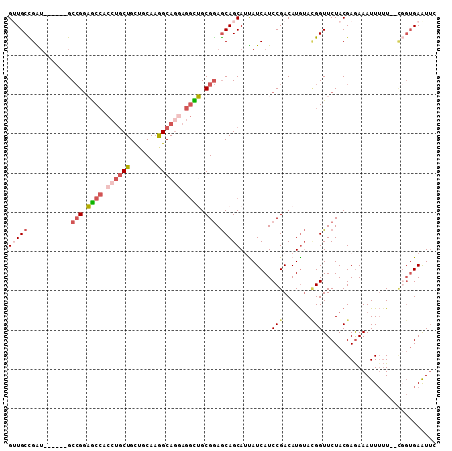
>3L_DroMel_CAF1 1616961 101 + 23771897 GUUGCCGAU------GCCGGAGCCACCUGCUGCUGCAAGGCAGGAGGCUGCGGAGCAGCAUUAUCAUCCGACAUGUACGGUUCUACGAGAAAUUUUU--CGGUGAGUUC (((((....------.(((.((((.((((((.......)))))).)))).))).)))))....(((((((.......))).....(((((....)))--)))))).... ( -37.10) >DroPse_CAF1 1831 103 + 1 GAUGCUGGU------GCCGGUACCACCUGCUGCUGCAAAGCAGGAGGUGGCGGAGCAGCAGCAUCAUCCGACAUGUACGGUUCUACGAGAAAUUUUUGUUCGUGAAUUG (((((((.(------(((.(..((((((.(((((....))))).))))))).).))).)))))))............((((((.(((((.........))))))))))) ( -45.90) >DroSec_CAF1 1688 101 + 1 GUUGCCGAU------GCCGGAGCCACCUGCUGCUGCAAGGCAGGAGGCUGCGGAGCAGCAUUAUCAUCCGACAUGUACGGUUCUACGAGAAAUUUUU--CGGUGAGUUC (((((....------.(((.((((.((((((.......)))))).)))).))).)))))....(((((((.......))).....(((((....)))--)))))).... ( -37.10) >DroSim_CAF1 1446 101 + 1 GUUGCCGAU------GCCGGAGCCACCUGCUGCUGCAAGGCAGGAGGCUGCGGAGCAGCAUUAUCAUCCGACAUGUACGGUUCUACGAGAAAUUUUU--CGGUGAGUUC (((((....------.(((.((((.((((((.......)))))).)))).))).)))))....(((((((.......))).....(((((....)))--)))))).... ( -37.10) >DroMoj_CAF1 1630 96 + 1 GUUGCUGGU------GCGGAUGCAUGAACCUGCUGCAAAGCAGGAGAUGGCGAAACAGCAGCAUCAUCCGACAUGAAAGGUUCUACGAAAAAUUUGU--G---GAAU-- .....(.((------(((((((.(((..((((((....)))))).....((......))..))))))))).))).)...(((((((((.....))))--)---))))-- ( -33.30) >DroAna_CAF1 1221 107 + 1 GAUGCCGGAGCCAGAGCCGGAGCCACCUGCUGUGGCACGGCAUUAGGCUGCGGAGCAGCAUUAUCAUCCGACAUAUAUGGUUCUACGAGAAAUUGUU--CGGUGAAUUG (((((((..((((.(((.((.....)).))).)))).)))))))..(((((...)))))....(((.((((((....)).....((((....)))))--)))))).... ( -41.10) >consensus GUUGCCGAU______GCCGGAGCCACCUGCUGCUGCAAGGCAGGAGGCUGCGGAGCAGCAUUAUCAUCCGACAUGUACGGUUCUACGAGAAAUUUUU__CGGUGAAUUC (((((...........(((.((((.((((((.......)))))).)))).))).)))))........(((.......)))............................. (-20.07 = -21.27 + 1.20)

| Location | 1,616,961 – 1,617,062 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 81.35 |
| Mean single sequence MFE | -32.76 |
| Consensus MFE | -18.61 |
| Energy contribution | -19.61 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.561158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
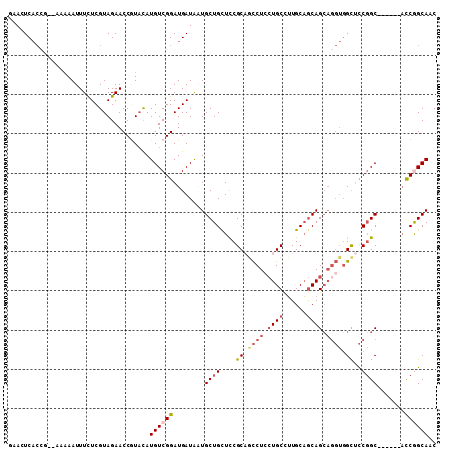
>3L_DroMel_CAF1 1616961 101 - 23771897 GAACUCACCG--AAAAAUUUCUCGUAGAACCGUACAUGUCGGAUGAUAAUGCUGCUCCGCAGCCUCCUGCCUUGCAGCAGCAGGUGGCUCCGGC------AUCGGCAAC ....((((((--(.........((......))......)))).)))...(((((..(((.((((.(((((.........))))).)))).))).------..))))).. ( -32.36) >DroPse_CAF1 1831 103 - 1 CAAUUCACGAACAAAAAUUUCUCGUAGAACCGUACAUGUCGGAUGAUGCUGCUGCUCCGCCACCUCCUGCUUUGCAGCAGCAGGUGGUACCGGC------ACCAGCAUC ...(((((((...........)))).)))(((.......)))..(((((((.((((..(((((((.(((((....))))).)))))))...)))------).))))))) ( -40.30) >DroSec_CAF1 1688 101 - 1 GAACUCACCG--AAAAAUUUCUCGUAGAACCGUACAUGUCGGAUGAUAAUGCUGCUCCGCAGCCUCCUGCCUUGCAGCAGCAGGUGGCUCCGGC------AUCGGCAAC ....((((((--(.........((......))......)))).)))...(((((..(((.((((.(((((.........))))).)))).))).------..))))).. ( -32.36) >DroSim_CAF1 1446 101 - 1 GAACUCACCG--AAAAAUUUCUCGUAGAACCGUACAUGUCGGAUGAUAAUGCUGCUCCGCAGCCUCCUGCCUUGCAGCAGCAGGUGGCUCCGGC------AUCGGCAAC ....((((((--(.........((......))......)))).)))...(((((..(((.((((.(((((.........))))).)))).))).------..))))).. ( -32.36) >DroMoj_CAF1 1630 96 - 1 --AUUC---C--ACAAAUUUUUCGUAGAACCUUUCAUGUCGGAUGAUGCUGCUGUUUCGCCAUCUCCUGCUUUGCAGCAGGUUCAUGCAUCCGC------ACCAGCAAC --....---.--........................((.((((((((((((((((...((........))...)))))))...))).)))))))------)........ ( -22.80) >DroAna_CAF1 1221 107 - 1 CAAUUCACCG--AACAAUUUCUCGUAGAACCAUAUAUGUCGGAUGAUAAUGCUGCUCCGCAGCCUAAUGCCGUGCCACAGCAGGUGGCUCCGGCUCUGGCUCCGGCAUC ...(((..((--(........)))..)))......((((((((......(((......)))(((....((((.(((((.....)))))..))))...))))))))))). ( -36.40) >consensus GAACUCACCG__AAAAAUUUCUCGUAGAACCGUACAUGUCGGAUGAUAAUGCUGCUCCGCAGCCUCCUGCCUUGCAGCAGCAGGUGGCUCCGGC______ACCGGCAAC ....................................((((((........((((....((.((((.((((......)))).)))).))..)))).......)))))).. (-18.61 = -19.61 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:25 2006