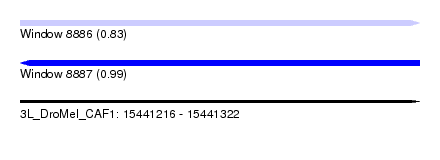
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,441,216 – 15,441,322 |
| Length | 106 |
| Max. P | 0.988383 |

| Location | 15,441,216 – 15,441,322 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 96.07 |
| Mean single sequence MFE | -20.33 |
| Consensus MFE | -18.56 |
| Energy contribution | -18.62 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15441216 106 + 23771897 UGAAAAUUUCGACGUAAAAAA-GUCGCGUCAUGUGCAAAAUUAUUGGGUAAUUAACGUGACACAGACACAACGGCAACGUCAAAGCAGACGAUGACUAUAAA--AAUAU ........(((.(((......-((((.(((((((....((((((...)))))).))))))).).)))...(((....)))........))).))).......--..... ( -20.30) >DroSec_CAF1 172512 106 + 1 UGAAAAUUUCGACGUAAAAAA-GUCGCGUCAUGUGCAAAAUUAUUGGGUAAUUAACGUGACACAGACACAACGGCAACGUCAAAGCAGACGAUGACUAUAAA--AAUAU ........(((.(((......-((((.(((((((....((((((...)))))).))))))).).)))...(((....)))........))).))).......--..... ( -20.30) >DroSim_CAF1 174255 106 + 1 UGAAAAUUUCGACGUAAAAAA-GUCGCGUCAUGUGCAAAAUUAUUGGGUAAUUAACGUGACACAGACACAACGGCAACGUCAAAGCAGACGAUGACUAUAAA--AAUAU ........(((.(((......-((((.(((((((....((((((...)))))).))))))).).)))...(((....)))........))).))).......--..... ( -20.30) >DroEre_CAF1 168980 106 + 1 UGAAAAUUUCGACGUAAAAAA-GUCGCGUCAUGUGCAAAAUUAUUGGGUAAUUAACGUGGCACAGACACAACGGCAACGUCACAGCAGACGAUGACUAUAAA--AAUAU ....................(-((((((((.(((((..((((((...))))))......)))))......(((....))).......)))).))))).....--..... ( -21.30) >DroYak_CAF1 174588 107 + 1 UGAAAAUUUCAACGUAAAAAACGUCGCGUCAUGUGCAAAAUUAUUGGGUAAUUAACGUGACACAGACACAACGGCAACGUCACAGCAGACGAUGACUAUAAA--AAUAU .....................(((((((((((((....((((((...)))))).))))))).........(((....)))....)).))))...........--..... ( -21.20) >DroAna_CAF1 310882 108 + 1 UGAAAAUUUCGCAGUAAAAAA-GUCGCGUCAUGUGCAAAAUUAUUGGGUAAUUAACGUGACACAGACACAACGGCAGCGACAAAGCAUACGAUGACUAUAAAAAAACAU ........((((.((......-((((.(((((((....((((((...)))))).))))))).).)))......)).))))............................. ( -18.60) >consensus UGAAAAUUUCGACGUAAAAAA_GUCGCGUCAUGUGCAAAAUUAUUGGGUAAUUAACGUGACACAGACACAACGGCAACGUCAAAGCAGACGAUGACUAUAAA__AAUAU ........(((.(((.......((((.(((((((....((((((...)))))).))))))).).)))...(((....)))........))).))).............. (-18.56 = -18.62 + 0.06)



| Location | 15,441,216 – 15,441,322 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 96.07 |
| Mean single sequence MFE | -23.95 |
| Consensus MFE | -21.60 |
| Energy contribution | -21.72 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15441216 106 - 23771897 AUAUU--UUUAUAGUCAUCGUCUGCUUUGACGUUGCCGUUGUGUCUGUGUCACGUUAAUUACCCAAUAAUUUUGCACAUGACGCGAC-UUUUUUACGUCGAAAUUUUCA .....--..................((((((((....(((((((((((((.(.((((.........)))).).))))).))))))))-......))))))))....... ( -24.40) >DroSec_CAF1 172512 106 - 1 AUAUU--UUUAUAGUCAUCGUCUGCUUUGACGUUGCCGUUGUGUCUGUGUCACGUUAAUUACCCAAUAAUUUUGCACAUGACGCGAC-UUUUUUACGUCGAAAUUUUCA .....--..................((((((((....(((((((((((((.(.((((.........)))).).))))).))))))))-......))))))))....... ( -24.40) >DroSim_CAF1 174255 106 - 1 AUAUU--UUUAUAGUCAUCGUCUGCUUUGACGUUGCCGUUGUGUCUGUGUCACGUUAAUUACCCAAUAAUUUUGCACAUGACGCGAC-UUUUUUACGUCGAAAUUUUCA .....--..................((((((((....(((((((((((((.(.((((.........)))).).))))).))))))))-......))))))))....... ( -24.40) >DroEre_CAF1 168980 106 - 1 AUAUU--UUUAUAGUCAUCGUCUGCUGUGACGUUGCCGUUGUGUCUGUGCCACGUUAAUUACCCAAUAAUUUUGCACAUGACGCGAC-UUUUUUACGUCGAAAUUUUCA .....--...(((((........)))))(((((....(((((((((((((...((((.........))))...))))).))))))))-......))))).......... ( -25.50) >DroYak_CAF1 174588 107 - 1 AUAUU--UUUAUAGUCAUCGUCUGCUGUGACGUUGCCGUUGUGUCUGUGUCACGUUAAUUACCCAAUAAUUUUGCACAUGACGCGACGUUUUUUACGUUGAAAUUUUCA ....(--((((..(((((.(....).))))).....((((((((((((((.(.((((.........)))).).))))).)))))))))..........)))))...... ( -23.60) >DroAna_CAF1 310882 108 - 1 AUGUUUUUUUAUAGUCAUCGUAUGCUUUGUCGCUGCCGUUGUGUCUGUGUCACGUUAAUUACCCAAUAAUUUUGCACAUGACGCGAC-UUUUUUACUGCGAAAUUUUCA ......(((..((((....(((.((......))))).(((((((((((((.(.((((.........)))).).))))).))))))))-......))))..)))...... ( -21.40) >consensus AUAUU__UUUAUAGUCAUCGUCUGCUUUGACGUUGCCGUUGUGUCUGUGUCACGUUAAUUACCCAAUAAUUUUGCACAUGACGCGAC_UUUUUUACGUCGAAAUUUUCA .........................((((((((....(((((((((((((...((((.........))))...))))).)))))))).......))))))))....... (-21.60 = -21.72 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:54 2006