
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,429,771 – 15,429,865 |
| Length | 94 |
| Max. P | 0.998966 |

| Location | 15,429,771 – 15,429,865 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 89.13 |
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -21.00 |
| Energy contribution | -22.20 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
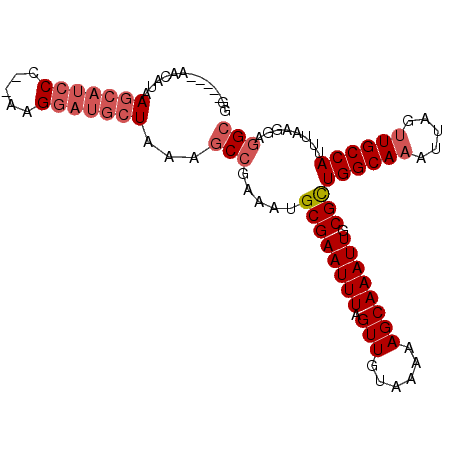
>3L_DroMel_CAF1 15429771 94 + 23771897 GCCUCCUUAAAUGGCAACUAAUUUGCCAGCGCAAUUUGCUUUUUACAACUAAAUUCGCAUUUCGGCUUUAGCAUCCUU--GGGAUGCUUAUGUU-----CC (((........((((((.....))))))(((.((((((.((.....)).))))))))).....)))...(((((((..--.)))))))......-----.. ( -25.10) >DroPse_CAF1 158792 95 + 1 UCUCCCUUUAAUGGCAACUAAU-UGCCAACGCAAUUUGCUUUUUAUAACUAAAUUCGCAAUUCGGCUUCAUUUUCCUCUUAGCUCUCUUUUCCU-----CC ...........((((((....)-)))))..((...((((..((((....))))...))))....))............................-----.. ( -12.10) >DroSec_CAF1 158727 94 + 1 GCCUCCUUAAAUGGCAACUAAUUUGCCAGCGCAAUUUGCUUUUUACAACUAAAUUCGCAUUUCGGCUUUAGCAUCCUU--GGGAUGCUUAUGCU-----CC (((........((((((.....))))))(((.((((((.((.....)).))))))))).....)))...(((((((..--.)))))))......-----.. ( -25.10) >DroSim_CAF1 163219 94 + 1 GCCUCCUUAAAUGGCAACUAAUUUGCCAGCGCAAUUUGCUUUUUACAACUAAAUUCGCAUUUCGGCUUUAGCAUCCUU--GGGAUGCUUAUGCU-----CC (((........((((((.....))))))(((.((((((.((.....)).))))))))).....)))...(((((((..--.)))))))......-----.. ( -25.10) >DroEre_CAF1 159511 94 + 1 GCCUUCUUAAAUGGCAACUAAUUUGCCAGCGCAAUUUGCUUUUUACAACUAAAUUCGCAUUUCGGCUUUAGCAUCCUU--GGGAUGCUUAUGUU-----CC (((........((((((.....))))))(((.((((((.((.....)).))))))))).....)))...(((((((..--.)))))))......-----.. ( -25.10) >DroYak_CAF1 164508 99 + 1 GCCUUCUUAAAUGGCAACUAAUUUGCCAGCGCAAUUUGCUUUUUACAACUAAAUUCGCAUUUUGGCUUUAGCAUCCUU--GGGAUGCUUAUGUUCCGUUCC (((........((((((.....))))))(((.((((((.((.....)).))))))))).....)))...(((((((..--.)))))))............. ( -24.80) >consensus GCCUCCUUAAAUGGCAACUAAUUUGCCAGCGCAAUUUGCUUUUUACAACUAAAUUCGCAUUUCGGCUUUAGCAUCCUU__GGGAUGCUUAUGCU_____CC (((........((((((.....))))))(((.((((((.((.....)).))))))))).....)))...(((((((.....)))))))............. (-21.00 = -22.20 + 1.20)



| Location | 15,429,771 – 15,429,865 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 89.13 |
| Mean single sequence MFE | -24.68 |
| Consensus MFE | -22.17 |
| Energy contribution | -23.37 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15429771 94 - 23771897 GG-----AACAUAAGCAUCCC--AAGGAUGCUAAAGCCGAAAUGCGAAUUUAGUUGUAAAAAGCAAAUUGCGCUGGCAAAUUAGUUGCCAUUUAAGGAGGC ..-----......(((((((.--..)))))))...(((.....((((((((.(((......)))))))).)))((((((.....))))))........))) ( -26.90) >DroPse_CAF1 158792 95 - 1 GG-----AGGAAAAGAGAGCUAAGAGGAAAAUGAAGCCGAAUUGCGAAUUUAGUUAUAAAAAGCAAAUUGCGUUGGCA-AUUAGUUGCCAUUAAAGGGAGA ..-----...........................((((((.((((...((((....))))..)))).))).)))((((-(....)))))............ ( -13.60) >DroSec_CAF1 158727 94 - 1 GG-----AGCAUAAGCAUCCC--AAGGAUGCUAAAGCCGAAAUGCGAAUUUAGUUGUAAAAAGCAAAUUGCGCUGGCAAAUUAGUUGCCAUUUAAGGAGGC ..-----......(((((((.--..)))))))...(((.....((((((((.(((......)))))))).)))((((((.....))))))........))) ( -26.90) >DroSim_CAF1 163219 94 - 1 GG-----AGCAUAAGCAUCCC--AAGGAUGCUAAAGCCGAAAUGCGAAUUUAGUUGUAAAAAGCAAAUUGCGCUGGCAAAUUAGUUGCCAUUUAAGGAGGC ..-----......(((((((.--..)))))))...(((.....((((((((.(((......)))))))).)))((((((.....))))))........))) ( -26.90) >DroEre_CAF1 159511 94 - 1 GG-----AACAUAAGCAUCCC--AAGGAUGCUAAAGCCGAAAUGCGAAUUUAGUUGUAAAAAGCAAAUUGCGCUGGCAAAUUAGUUGCCAUUUAAGAAGGC ..-----......(((((((.--..)))))))...(((.....((((((((.(((......)))))))).)))((((((.....))))))........))) ( -26.90) >DroYak_CAF1 164508 99 - 1 GGAACGGAACAUAAGCAUCCC--AAGGAUGCUAAAGCCAAAAUGCGAAUUUAGUUGUAAAAAGCAAAUUGCGCUGGCAAAUUAGUUGCCAUUUAAGAAGGC .............(((((((.--..)))))))...(((.....((((((((.(((......)))))))).)))((((((.....))))))........))) ( -26.90) >consensus GG_____AACAUAAGCAUCCC__AAGGAUGCUAAAGCCGAAAUGCGAAUUUAGUUGUAAAAAGCAAAUUGCGCUGGCAAAUUAGUUGCCAUUUAAGGAGGC .............(((((((.....)))))))...(((.....((((((((.(((......)))))))).)))((((((.....))))))........))) (-22.17 = -23.37 + 1.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:51 2006