| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,397,920 – 15,398,014 |
| Length | 94 |
| Max. P | 0.983066 |

| Location | 15,397,920 – 15,398,014 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 77.65 |
| Mean single sequence MFE | -22.87 |
| Consensus MFE | -15.04 |
| Energy contribution | -16.35 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.983066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15397920 94 + 23771897 AUUUCU------AGUUUUUAUUUUUUUUCAUUCUCUUUUCCCACUUUCGGUGUCUCGAUCAGAUACCAAAAGGAUGGCCUCGGAAAAAAUAGAGUGCAGA ......------................((((((.((((((..((((.(((((((.....))))))).))))((.....))))))))...)))))).... ( -20.20) >DroSec_CAF1 126832 94 + 1 AUUUCU------AGUUUUUAUUUUUUUUCAUUCUCUUUUCCCACUUUCGGUGUCUCGAUCAGAUACCAAAAGGAUGGCCUCGGAAAAAAUAGAGUGCAGA ......------................((((((.((((((..((((.(((((((.....))))))).))))((.....))))))))...)))))).... ( -20.20) >DroSim_CAF1 129098 94 + 1 NNNNNN------NNNNNNNNNNNNNNUUCAUUCUCUUUUCCCACUUUCGGUGUCUCGUUUAGAUACCGAAAGGAUGGCCUCGGAAAAAAUAGAGUGCAGA ......------................((((((.((((((..((((((((((((.....))))))))))))((.....))))))))...)))))).... ( -26.00) >DroEre_CAF1 127651 90 + 1 UUCUCU------AGUUUUUAU---UUUUCAUUCUGUUUUCCCACUUUCGGUGUCUCGAUCAGAUACCA-AAGGAUGGCCUCGGAAAAAAUAGAGAGCAGA ...(((------..(((((((---((((...((((.....(((((((.(((((((.....))))))).-)))).)))...)))))))))))))))..))) ( -24.10) >DroYak_CAF1 130837 90 + 1 UUUUCU------AGUUUUUAU---UUUUCAUUCUGUUUUCCCACUUUCGGUGUCUCGAUCAGAUACCA-AAGGAUGGCCUCGGAAAAAAUAGAGAGCAGA ...(((------..(((((((---((((...((((.....(((((((.(((((((.....))))))).-)))).)))...)))))))))))))))..))) ( -24.20) >DroAna_CAF1 272354 89 + 1 UUUUUUUUUGACAUUUUUUUUUUUUGUCCGUUCUAUUUUUCCACUUUUGGUGUCUCAAUCAGAUACCAAACGGAUGGCUU-----------GAAUGGGGA ..........................(((((((......(((...((((((((((.....)))))))))).)))......-----------))))))).. ( -22.50) >consensus UUUUCU______AGUUUUUAU___UUUUCAUUCUCUUUUCCCACUUUCGGUGUCUCGAUCAGAUACCAAAAGGAUGGCCUCGGAAAAAAUAGAGUGCAGA ............................((((((.((((((.......(((((((.....)))))))...(((....))).))))))...)))))).... (-15.04 = -16.35 + 1.31)

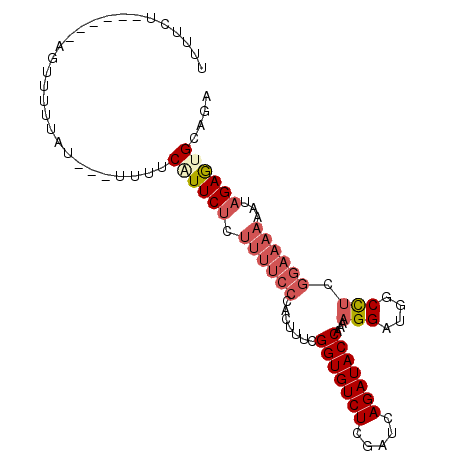
| Location | 15,397,920 – 15,398,014 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 77.65 |
| Mean single sequence MFE | -18.50 |
| Consensus MFE | -12.70 |
| Energy contribution | -12.98 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15397920 94 - 23771897 UCUGCACUCUAUUUUUUCCGAGGCCAUCCUUUUGGUAUCUGAUCGAGACACCGAAAGUGGGAAAAGAGAAUGAAAAAAAAUAAAAACU------AGAAAU ....((.(((.((((((((((.....))((((((((.(((.....))).)))))))).))))))))))).))................------...... ( -20.10) >DroSec_CAF1 126832 94 - 1 UCUGCACUCUAUUUUUUCCGAGGCCAUCCUUUUGGUAUCUGAUCGAGACACCGAAAGUGGGAAAAGAGAAUGAAAAAAAAUAAAAACU------AGAAAU ....((.(((.((((((((((.....))((((((((.(((.....))).)))))))).))))))))))).))................------...... ( -20.10) >DroSim_CAF1 129098 94 - 1 UCUGCACUCUAUUUUUUCCGAGGCCAUCCUUUCGGUAUCUAAACGAGACACCGAAAGUGGGAAAAGAGAAUGAANNNNNNNNNNNNNN------NNNNNN ....((.(((.((((((((((.....))((((((((.(((.....))).)))))))).))))))))))).))................------...... ( -22.10) >DroEre_CAF1 127651 90 - 1 UCUGCUCUCUAUUUUUUCCGAGGCCAUCCUU-UGGUAUCUGAUCGAGACACCGAAAGUGGGAAAACAGAAUGAAAA---AUAAAAACU------AGAGAA .....((((((.(((((......((((..((-((((.(((.....))).)))))).))))..........(....)---..))))).)------))))). ( -17.80) >DroYak_CAF1 130837 90 - 1 UCUGCUCUCUAUUUUUUCCGAGGCCAUCCUU-UGGUAUCUGAUCGAGACACCGAAAGUGGGAAAACAGAAUGAAAA---AUAAAAACU------AGAAAA ((((.((((..........))))((((..((-((((.(((.....))).)))))).)))).....)))).......---.........------...... ( -16.30) >DroAna_CAF1 272354 89 - 1 UCCCCAUUC-----------AAGCCAUCCGUUUGGUAUCUGAUUGAGACACCAAAAGUGGAAAAAUAGAACGGACAAAAAAAAAAAAUGUCAAAAAAAAA ...((.(((-----------...((((...((((((.(((.....))).)))))).)))).......))).))........................... ( -14.60) >consensus UCUGCACUCUAUUUUUUCCGAGGCCAUCCUUUUGGUAUCUGAUCGAGACACCGAAAGUGGGAAAAGAGAAUGAAAA___AUAAAAACU______AGAAAA (((...(((..........))).(((..((((((((.(((.....))).)))))))))))......)))............................... (-12.70 = -12.98 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:39 2006