| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,381,130 – 15,381,220 |
| Length | 90 |
| Max. P | 0.941484 |

| Location | 15,381,130 – 15,381,220 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 87.09 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -19.60 |
| Energy contribution | -20.35 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
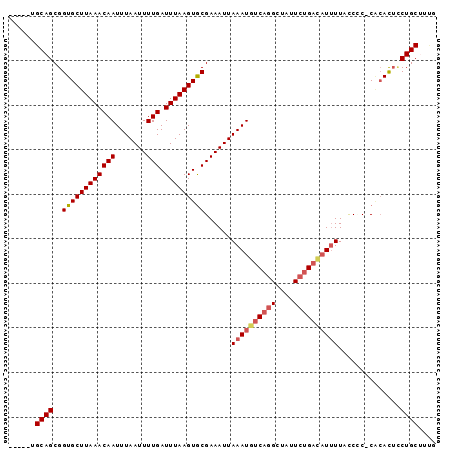
>3L_DroMel_CAF1 15381130 90 + 23771897 -----UGCAGCGGUGCUUAAACAAUUUAAUUUUGAUUUAAGUGCGAAAUUAAAUGUCAGGCUAUUCUGAAAUUUUAUCCC-CACACUGCUGCUUUG -----.(((((((((........(((((((((((((....)).))))))))))).(((((....)))))...........-..))))))))).... ( -25.70) >DroSec_CAF1 112891 90 + 1 -----UGCAGCGGUGCUUAAACAAUUUAAUUUUGAUUUAAGUGCGAAAUUAAAUGUCAGGCUAUUCUGACAUUUUACCCC-CACACUCCUGCUUUG -----.((((.((..(((((((((.......))).))))))..)......((((((((((....))))))))))......-......))))).... ( -24.10) >DroSim_CAF1 115233 90 + 1 -----UGCAGCGGUGCUUAAACAAUUUAAUUUUGAUUUAAGUGCGAAAUUAAAUGUCAGGCUAUUCUGACAUUUUACCCC-CACACUCCUGCUUUG -----.((((.((..(((((((((.......))).))))))..)......((((((((((....))))))))))......-......))))).... ( -24.10) >DroEre_CAF1 113729 88 + 1 -----UGCAGCGGUGCUUAAACAAUUUAAUUUUGAUUUAAGUGCGAAAUUAAAUGUCAGGCCAUUCUGACAUUUU-CC-C-CGCACUCCUGCCUCC -----.((((.(((((((((((((.......))).)))))))(((.....((((((((((....)))))))))).-..-.-)))))).)))).... ( -27.60) >DroYak_CAF1 116300 89 + 1 -----UGCAGCGGUGCUUAAACAAUUUAAUUUUGAUUUAAGUGCGAAAUUAAAUGUCAGGCUAUUCUGACAUUUU-CCAC-CACACUCCUGCCUCG -----.((((.((..(((((((((.......))).))))))..)......((((((((((....)))))))))).-....-......))))).... ( -23.70) >DroAna_CAF1 259455 94 + 1 GCAGCUGCAGCGGUGCUUAAACAAUUUAAUUUUGAUUUAAGUACGAAAUUAAAUGUCAGGAAAAUGCGCUUUCUUUCCUCUCUAAUAUCUGCU--G .......(((((((((((((((((.......))).)))))))))((.((((...(..((((((..........))))))..))))).))))))--) ( -19.40) >consensus _____UGCAGCGGUGCUUAAACAAUUUAAUUUUGAUUUAAGUGCGAAAUUAAAUGUCAGGCUAUUCUGACAUUUUACCCC_CACACUCCUGCUUUG ......((((..((((((((((((.......))).)))))))))......((((((((((....))))))))))..............)))).... (-19.60 = -20.35 + 0.75)


| Location | 15,381,130 – 15,381,220 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 87.09 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -17.90 |
| Energy contribution | -19.32 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
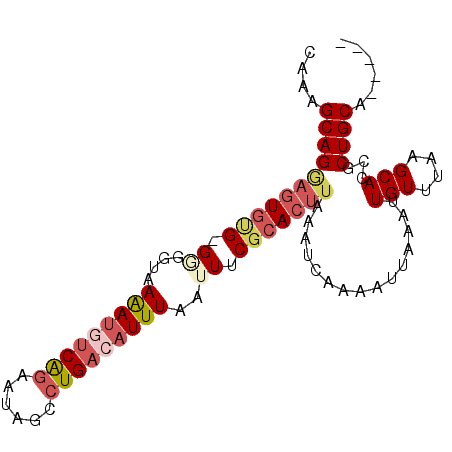
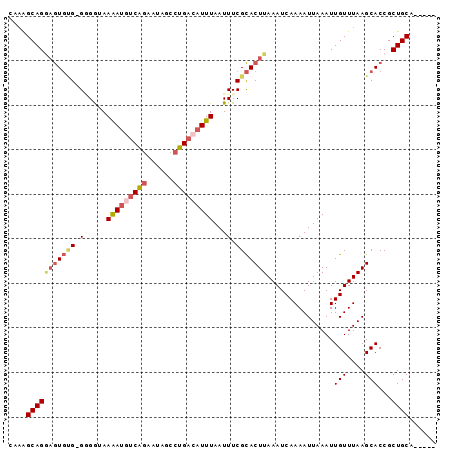
>3L_DroMel_CAF1 15381130 90 - 23771897 CAAAGCAGCAGUGUG-GGGAUAAAAUUUCAGAAUAGCCUGACAUUUAAUUUCGCACUUAAAUCAAAAUUAAAUUGUUUAAGCACCGCUGCA----- ....(((((.(((((-(..((.((((.((((......)))).)))).))..).))((((((.(((.......)))))))))))).))))).----- ( -24.40) >DroSec_CAF1 112891 90 - 1 CAAAGCAGGAGUGUG-GGGGUAAAAUGUCAGAAUAGCCUGACAUUUAAUUUCGCACUUAAAUCAAAAUUAAAUUGUUUAAGCACCGCUGCA----- ....(((((((((((-..(...(((((((((......)))))))))..)..)))))))...............(((....)))...)))).----- ( -29.20) >DroSim_CAF1 115233 90 - 1 CAAAGCAGGAGUGUG-GGGGUAAAAUGUCAGAAUAGCCUGACAUUUAAUUUCGCACUUAAAUCAAAAUUAAAUUGUUUAAGCACCGCUGCA----- ....(((((((((((-..(...(((((((((......)))))))))..)..)))))))...............(((....)))...)))).----- ( -29.20) >DroEre_CAF1 113729 88 - 1 GGAGGCAGGAGUGCG-G-GG-AAAAUGUCAGAAUGGCCUGACAUUUAAUUUCGCACUUAAAUCAAAAUUAAAUUGUUUAAGCACCGCUGCA----- ....(((((((((((-.-.(-.(((((((((......)))))))))..)..)))))))...............(((....)))...)))).----- ( -29.60) >DroYak_CAF1 116300 89 - 1 CGAGGCAGGAGUGUG-GUGG-AAAAUGUCAGAAUAGCCUGACAUUUAAUUUCGCACUUAAAUCAAAAUUAAAUUGUUUAAGCACCGCUGCA----- ....(((((((((((-(...-.(((((((((......)))))))))....))))))))...............(((....)))...)))).----- ( -26.60) >DroAna_CAF1 259455 94 - 1 C--AGCAGAUAUUAGAGAGGAAAGAAAGCGCAUUUUCCUGACAUUUAAUUUCGUACUUAAAUCAAAAUUAAAUUGUUUAAGCACCGCUGCAGCUGC (--(((.((.((((((.(((((((........)))))))....)))))).))((.((((((.(((.......))))))))).)).))))....... ( -18.30) >consensus CAAAGCAGGAGUGUG_GGGGUAAAAUGUCAGAAUAGCCUGACAUUUAAUUUCGCACUUAAAUCAAAAUUAAAUUGUUUAAGCACCGCUGCA_____ ....(((((((((((.((....(((((((((......)))))))))..)).)))))))...............(((....)))...))))...... (-17.90 = -19.32 + 1.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:33 2006