
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,367,220 – 15,367,320 |
| Length | 100 |
| Max. P | 0.886367 |

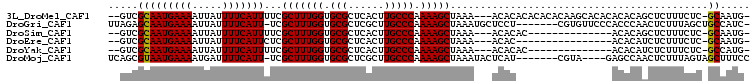
| Location | 15,367,220 – 15,367,320 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.58 |
| Mean single sequence MFE | -17.15 |
| Consensus MFE | -14.24 |
| Energy contribution | -14.10 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
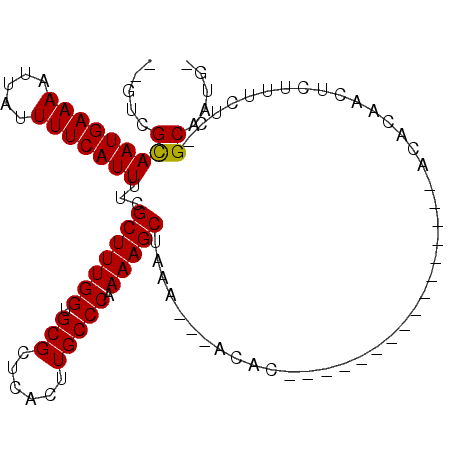
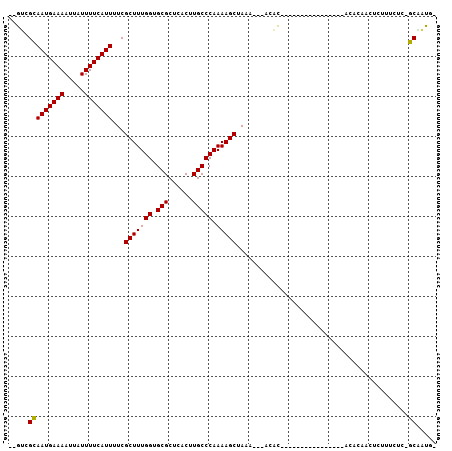
>3L_DroMel_CAF1 15367220 100 + 23771897 --GUCGCAAUGAAAAUUAUUUUCAUUUUCGCUUUGGUGCGCUCACUUGCCCAAAAGCUAAA---ACACACACACACAAGCACACACACAGCUCUUUCUC-GCAAUG- --((.(((((((((.....)))))))...(((((((.(((......))))).)))))....---..............)))).......((........-))....- ( -16.40) >DroGri_CAF1 97725 98 + 1 UUAGAGCAAUGAAAAUUAUUUUCAUU-UCGCUUUGGUGCGCUCGCUUGCCCAAAAGCUAAAUGCUCCU-------CGUGUUCCCACCCAACUCUUUAGCUGCCAUC- ...(((((((((((.....)))))).-..(((((((.(((......))))).)))))....)))))..-------.(((....)))....................- ( -19.30) >DroSim_CAF1 101001 86 + 1 --GUCGCAAUGAAAAUUAUUUUCAUUUUCGCUUUGGUGCGCUCACUUGCCCAAAAGCUAAA---ACACAC--------------ACACAGCUCUUUCUC-GCAAUG- --...(((((((((.....)))))))...(((((((.(((......))))).)))))....---......--------------...............-))....- ( -15.20) >DroEre_CAF1 98744 84 + 1 --GUCGCAAUGAAAAUUAUUUUCAUUCUCGCUUUGGUGCGCUCACUUGCCCAAAAGCUAAA---ACAC----------------ACACAUCUCUUUCUC-GCAAUG- --...(((((((((.....)))))))...(((((((.(((......))))).)))))....---....----------------...............-))....- ( -15.20) >DroYak_CAF1 102051 86 + 1 --GUCGCAAUGAAAAUUAUUUUCAUUUUCGCUUUGGUGCGCUCACUUGCCCAAAAGCUAAA---ACACAC--------------ACACAUCUCUUUCUC-GCCAUG- --...(((((((((.....)))))))...(((((((.(((......))))).)))))....---......--------------...............-))....- ( -14.30) >DroMoj_CAF1 95656 95 + 1 UCAGCGUAAUGAAAAUGAUUUUCAUU-UCGCUUUGGUGCGCUCGCUUGCCCAAAAGCUAAAUACUCAU-------CGUA----GAGCCAACUCUUUAGUAGCUUUCC ..((((.(((((((.....)))))))-.))))((((.(((......)))))))((((((..(((....-------.)))----(((....))).....))))))... ( -22.50) >consensus __GUCGCAAUGAAAAUUAUUUUCAUUUUCGCUUUGGUGCGCUCACUUGCCCAAAAGCUAAA___ACAC________________ACACAACUCUUUCUC_GCAAUG_ .....(((((((((.....)))))))...(((((((.(((......))))).)))))...........................................))..... (-14.24 = -14.10 + -0.14)
| Location | 15,367,220 – 15,367,320 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.58 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -13.02 |
| Energy contribution | -13.02 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
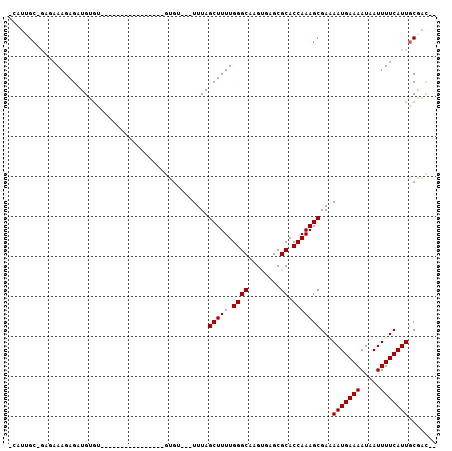
>3L_DroMel_CAF1 15367220 100 - 23771897 -CAUUGC-GAGAAAGAGCUGUGUGUGUGCUUGUGUGUGUGUGU---UUUAGCUUUUGGGCAAGUGAGCGCACCAAAGCGAAAAUGAAAAUAAUUUUCAUUGCGAC-- -..((((-((......(((.((.(((((((..(.(((.((.((---....))...)).))).)..))))))))).)))((((((.......)))))).)))))).-- ( -30.80) >DroGri_CAF1 97725 98 - 1 -GAUGGCAGCUAAAGAGUUGGGUGGGAACACG-------AGGAGCAUUUAGCUUUUGGGCAAGCGAGCGCACCAAAGCGA-AAUGAAAAUAAUUUUCAUUGCUCUAA -.....(((((....))))).(((....))).-------.(((((.....(((((.((((........)).)))))))..-(((((((.....)))))))))))).. ( -28.80) >DroSim_CAF1 101001 86 - 1 -CAUUGC-GAGAAAGAGCUGUGU--------------GUGUGU---UUUAGCUUUUGGGCAAGUGAGCGCACCAAAGCGAAAAUGAAAAUAAUUUUCAUUGCGAC-- -..((((-((......(((.((.--------------((((((---((..(((....)))....)))))))))).)))((((((.......)))))).)))))).-- ( -26.20) >DroEre_CAF1 98744 84 - 1 -CAUUGC-GAGAAAGAGAUGUGU----------------GUGU---UUUAGCUUUUGGGCAAGUGAGCGCACCAAAGCGAGAAUGAAAAUAAUUUUCAUUGCGAC-- -..((((-((........((.((----------------((((---((..(((....)))....))))))))))....((((((.......)))))).)))))).-- ( -21.20) >DroYak_CAF1 102051 86 - 1 -CAUGGC-GAGAAAGAGAUGUGU--------------GUGUGU---UUUAGCUUUUGGGCAAGUGAGCGCACCAAAGCGAAAAUGAAAAUAAUUUUCAUUGCGAC-- -....((-((......(...((.--------------((((((---((..(((....)))....))))))))))...)((((((.......)))))).))))...-- ( -20.60) >DroMoj_CAF1 95656 95 - 1 GGAAAGCUACUAAAGAGUUGGCUC----UACG-------AUGAGUAUUUAGCUUUUGGGCAAGCGAGCGCACCAAAGCGA-AAUGAAAAUCAUUUUCAUUACGCUGA ((...(((....((((((((((((----....-------..))))...)))))))).)))..((....)).))..((((.-(((((((.....))))))).)))).. ( -27.30) >consensus _CAUUGC_GAGAAAGAGAUGUGU________________GUGU___UUUAGCUUUUGGGCAAGUGAGCGCACCAAAGCGAAAAUGAAAAUAAUUUUCAUUGCGAC__ ..................................................(((((.((((........)).)))))))...(((((((.....)))))))....... (-13.02 = -13.02 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:25 2006